Team:NYMU-Taipei/modeling/m4
From 2014.igem.org
| Line 2: | Line 2: | ||
<html> | <html> | ||
<div id="main-content"> | <div id="main-content"> | ||
| - | <h> | + | <h>Stephen curve model</h> |
| - | <h1> | + | <h1>Purpose</h1> |
| - | <p>< | + | <p>1. To quantize and prove the effectiveness of our product on “Stephan curve”, a crucial curve for dental decay assessment which shows pH drop in mouth after ingestion.<br> 2. To figure out the effective duration of our product and display it in term of Stephan curve.</p> |
| - | + | <h1>Introduction</h1> | |
| - | <h1> | + | <p>Stephan curve illustrates the change in oral pH within few minutes in response to a cariogenic challenge, which is mainly caused by the bacterial metabolism of sugar-rich food. As we have mentioned, oral pH plays important role in caries assessment, since the solubility and demineralization[1] of the tooth is pH dependent[2]. The longer time oral pH is below “critical pH 5.5”, the more likely dental plaque is to take place. Therefore, the cariogenicity is measured as the area delimited by the Critical pH and the Stephan Curve[3].<br><br> |
| - | <h1>result</h1> | + | In our <a href='/Team:NYMU-Taipei/modeling/m2'><b>competition model</b></a>, we have shown how the population shift would cause the change of pH value. As population shift in oral cavity is a long term process, the model we built and the data we fit in competition model are in the view of large time scale (few days). Therefore, the model only provides information about how our product affects the final pH result from bacterial metabolism, that is, the minimum pH value in Stephan curve. However, dentistry experts gave us suggestions that besides from the raising of minimum pH, showing how our product influence Stephan curve to dentists and the public may be more persuasive.<br> |
| - | <h1> | + | Here we try to use the bacteria population derived in competition model to fit the general Stephan curve proposed by RM Stephan[4]. Studies have shown that in short time scale, the change of pH is proportional to the amount of bacteria, which correlates to our growth & pH model. Moreover, there are also some driving forces such as salivary[5], water, or oxygen content that would buffer and rebound the pH value. Therefore, we build a model for Stephan curve simulation with the consideration of bacteria amount and a upward driving force.<br><br> |
| + | Although the aim of our project is to provide a long term oral care for everyone, there are still some concerns about the effective duration of our product. Both the flux of salivary and drinks would reduce the amount of phage we apply in oral cavity, as well as the M102 phage resistance result from selection. Therefore, we do literature search to figure out the probability of phage loss [6] and S. mutans resistance, and estimate the effective duration of our product via the change of Stephan curve. | ||
| + | </p> | ||
| + | <h1>Models and mathematic equations</h1> | ||
| + | <p>Three well known growth models (Logistic, Gompertz, and Richartz)are used in this work. Characteristic model parameters (such as lag phase (λ), maximal growth rate (µ-max slope), stationary phase (A-max growth value)) are derived from our experimental data. Bootstrap and cross-validation techniques are used for estimating confidence intervals of | ||
| + | all derived parameters.<br><br> | ||
| + | The aim is to integrate the experimental data into different growth models and to compare the models using statistical methods (AIC and maximum likelihood were used). We believe, and many scientists do, that model selection is the most important part in model-experiment based research-<i>The right data with the right model</i>.</p><br> | ||
| + | <p> | ||
| + | <b>Logistic Model:</b> | ||
| + | $$\begin{align} | ||
| + | y(t)&=\frac{A}{1+\exp\left(\frac{4\mu}{A}(\lambda-t)+2\right)} | ||
| + | \end{align}$$ | ||
| + | <b>Gompertz Model:</b> | ||
| + | $$\begin{align} | ||
| + | y(t)&=A.\exp\left[-\exp\left(\frac{\mu.\exp(1)}{A}(\lambda-t)+1\right)\right] | ||
| + | \end{align}$$ | ||
| + | <b>Richartz Model:</b> | ||
| + | \begin{align} | ||
| + | y(t)&=A.\left[1+\nu\exp\left(1+\nu+\frac{\mu}{A}(1+\nu)^{1+1/\nu}(\lambda-t)\right)\right]^{-1/\nu} | ||
| + | \end{align} | ||
| + | $\nu$ is a shape parameter (in the richartz model only) | ||
| + | </p> | ||
| + | <h1>pH model</h1> | ||
| + | <p>Existed model is | ||
| + | $$\begin{align} | ||
| + | \frac{dpH}{dP}&=k(pH-pH_{min}) | ||
| + | \end{align}$$ | ||
| + | However, this model is not appropriate for our experimental data; we used linear regression model instead. | ||
| + | $$pH=\alpha + \beta OD +\epsilon$$ | ||
| + | where $\epsilon$ is a random error or noice (which helps to capture a measurement error and other unknown factors). And it is assumed to be Gaussian (normal distribution function), $\epsilon = N(\theta,\sigma^{2})$, with mean $\theta$ and constant variance $\sigma^{2}$. $\alpha$ and $\beta$ are model parameters. | ||
| + | </p> | ||
| + | <h1>Result and model validation</h1> | ||
| + | <img src="/wiki/images/a/af/NYMU14_model1_simulGrowth.png" style="display: block;width: 500px;margin: 0 auto;"><p style=" text-align: center; ">Figure 1: Simulation results of the growth curve</p><br> | ||
| + | <p>Our experimental data are implemented in the three proposed models. AIC is used to measure the performance of the models and the result shows that Logistic and Richartz models are approximately the same, but slightly different from Gompertz. However, using 95% confidence interval all of them are appropriate to fit the given data.<br><br> | ||
| + | <img src="/wiki/images/5/52/NYMU14_model1_modelValidation.png" style="display: block;width: 500px;margin: 0 auto;"><p style=" text-align: center; ">Figure 2: Shows validation test(t*) for the fitted model</p><br> | ||
| + | <p>The validation test (plot of the residuals), as Figure 2, shows that the simulation result obtained from bootstrap samples is suitable to estimate the model parameters.<br><br> | ||
| + | Using our experimental data as initials samples, we applied Bootsrap statistical to sample empirical data. The simulated result is presented as Figure (1). And, the estimated model parameters (mu(µ), lamda(λ), A) are summarized as Table 1. | ||
| + | $$\text{Table 1: Estimated Model Parameter}$$ | ||
| + | </p> | ||
| + | <pre> | ||
| + | ------------------------------------------- | ||
| + | mu lamda A | ||
| + | =========================================== | ||
| + | Lower 0.294 16.041 1.155 | ||
| + | Mean 0.384 16.456 1.187 | ||
| + | Upper 0.474 16.871 1.220 | ||
| + | Std 0.046 0.212 0.016 | ||
| + | ------------------------------------------- | ||
| + | Integral value: 16.622 | ||
| + | ------------------------------------------- | ||
| + | </pre> | ||
| + | <p> | ||
| + | From the result on table $1$, the $lower$ and $upper$ values are the estimated confidence intervals of the corresponding parameters. $Std$-standard deviation) and $mean$-average values are obtained from the bootstrap samples. These results are used in the proposed three growth models. After the approprate models are selected (in our case, the three proposed models are equally approprate), the parameters are taken as an intial values for the interaction model; which is used to study the growth of S.mutant in the presence of other species(see <a href='/Team:NYMU-Taipei/modeling/m2'><b>competition model</b></a>). | ||
| + | </p> | ||
| + | <p>$$\text{Table 2: Analysis of Regression Model}$$</p> | ||
| + | <pre> | ||
| + | ============================================================== | ||
| + | Estimated parametrs: | Statistical Tests: | ||
| + | Intercept (alpha)=7.6074 | R-squared= 0.9135 | ||
| + | OD (beta)=-2.0365 | p-value=6.228e-14 | ||
| + | -------------------------------------------------------------- | ||
| + | F-statistic=254.6 | ||
| + | =============================================================== | ||
| + | </pre> | ||
| + | <p>Using the table 2 result, the pH model can be estimated as: | ||
| + | $$pH=7.61 - 2.04 OD$$ | ||
| + | There is a strong linear correlation between pH and OD (R=0.9135, F=254.5, P.value $<$0.05). The coefficient value indicates that for every additional unit in OD we can expect pH to decrease by an average of 2.04. For examples: if the OD=1, pH is expected to be (pH=7.61 - 2.04(1))=5.57. The red fitted line graphically shows the same information.</p><br> | ||
| + | <img src='/wiki/images/2/25/NYMU14_model1_FittedModels.png' style="display: block;width: 700px;margin: 0 auto;"><p style=" text-align: center; ">Figure 3: Shows model comparison-the shaded regions indicate a 95% confiden interval of the model fits (bold lines)</p><br> | ||
| + | <p>If we move left or right along the x-axis by an amount that represents a one unit change in OD, the fitted line falls by 2.04 units. If the fitted line was flat (a slope coefficient of zero), the expected value for pH would not change no matter how far up and down the line you go. So, the low p-value ($\leq$ 0.05) suggests that the slope is not zero, which in turn suggests that changes in the predictor variable(OD) are associated with changes in the response variable (pH). <i>R-squared</i> is a statistical measure of how close the data are to the fitted regression line. It is also known as the coefficient of determination. It is the percentage of the response variable variation that is explained by a linear model. In this case, R-square =0.9135 shows that 91.35% of the pH variation is explained by OD, and the remain 8.65 can be explained by other factors that are not considered in the regression model. </p> | ||
| + | <img src="/wiki/images/6/61/NYMU14_model1_ciODph.png" style="display: block;width: 500px;margin: 0 auto;"><p style=" text-align: center; ">Figure 4: ph vs OD with 95% confident interval</p><br> | ||
| + | <img src="/wiki/images/1/11/NYMU14_model1_regression.png" style="display: block;width: 500px;margin: 0 auto;"><p style=" text-align: center; ">Figure 5: Model Fit</p><br> | ||
| + | <img src="/wiki/images/4/43/NYMU14_model1_residuals.png" style="display: block;width: 500px;margin: 0 auto;"><p style=" text-align: center; ">Figure 6: Validation Test</p><br> | ||
| + | <h1>Conclusion</h1> | ||
| + | <h1>Reference</h1> | ||
| + | |||
</div> | </div> | ||
</html> | </html> | ||
| + | {{:Team:NYMU-Taipei/NYMU14_Footer}} | ||
Revision as of 06:25, 27 September 2014
Purpose
1. To quantize and prove the effectiveness of our product on “Stephan curve”, a crucial curve for dental decay assessment which shows pH drop in mouth after ingestion.
2. To figure out the effective duration of our product and display it in term of Stephan curve.
Introduction
Stephan curve illustrates the change in oral pH within few minutes in response to a cariogenic challenge, which is mainly caused by the bacterial metabolism of sugar-rich food. As we have mentioned, oral pH plays important role in caries assessment, since the solubility and demineralization[1] of the tooth is pH dependent[2]. The longer time oral pH is below “critical pH 5.5”, the more likely dental plaque is to take place. Therefore, the cariogenicity is measured as the area delimited by the Critical pH and the Stephan Curve[3].
In our competition model, we have shown how the population shift would cause the change of pH value. As population shift in oral cavity is a long term process, the model we built and the data we fit in competition model are in the view of large time scale (few days). Therefore, the model only provides information about how our product affects the final pH result from bacterial metabolism, that is, the minimum pH value in Stephan curve. However, dentistry experts gave us suggestions that besides from the raising of minimum pH, showing how our product influence Stephan curve to dentists and the public may be more persuasive.
Here we try to use the bacteria population derived in competition model to fit the general Stephan curve proposed by RM Stephan[4]. Studies have shown that in short time scale, the change of pH is proportional to the amount of bacteria, which correlates to our growth & pH model. Moreover, there are also some driving forces such as salivary[5], water, or oxygen content that would buffer and rebound the pH value. Therefore, we build a model for Stephan curve simulation with the consideration of bacteria amount and a upward driving force.
Although the aim of our project is to provide a long term oral care for everyone, there are still some concerns about the effective duration of our product. Both the flux of salivary and drinks would reduce the amount of phage we apply in oral cavity, as well as the M102 phage resistance result from selection. Therefore, we do literature search to figure out the probability of phage loss [6] and S. mutans resistance, and estimate the effective duration of our product via the change of Stephan curve.
Models and mathematic equations
Three well known growth models (Logistic, Gompertz, and Richartz)are used in this work. Characteristic model parameters (such as lag phase (λ), maximal growth rate (µ-max slope), stationary phase (A-max growth value)) are derived from our experimental data. Bootstrap and cross-validation techniques are used for estimating confidence intervals of
all derived parameters.
The aim is to integrate the experimental data into different growth models and to compare the models using statistical methods (AIC and maximum likelihood were used). We believe, and many scientists do, that model selection is the most important part in model-experiment based research-The right data with the right model.
Logistic Model: $$\begin{align} y(t)&=\frac{A}{1+\exp\left(\frac{4\mu}{A}(\lambda-t)+2\right)} \end{align}$$ Gompertz Model: $$\begin{align} y(t)&=A.\exp\left[-\exp\left(\frac{\mu.\exp(1)}{A}(\lambda-t)+1\right)\right] \end{align}$$ Richartz Model: \begin{align} y(t)&=A.\left[1+\nu\exp\left(1+\nu+\frac{\mu}{A}(1+\nu)^{1+1/\nu}(\lambda-t)\right)\right]^{-1/\nu} \end{align} $\nu$ is a shape parameter (in the richartz model only)
pH model
Existed model is $$\begin{align} \frac{dpH}{dP}&=k(pH-pH_{min}) \end{align}$$ However, this model is not appropriate for our experimental data; we used linear regression model instead. $$pH=\alpha + \beta OD +\epsilon$$ where $\epsilon$ is a random error or noice (which helps to capture a measurement error and other unknown factors). And it is assumed to be Gaussian (normal distribution function), $\epsilon = N(\theta,\sigma^{2})$, with mean $\theta$ and constant variance $\sigma^{2}$. $\alpha$ and $\beta$ are model parameters.
Result and model validation

Figure 1: Simulation results of the growth curve
Our experimental data are implemented in the three proposed models. AIC is used to measure the performance of the models and the result shows that Logistic and Richartz models are approximately the same, but slightly different from Gompertz. However, using 95% confidence interval all of them are appropriate to fit the given data.
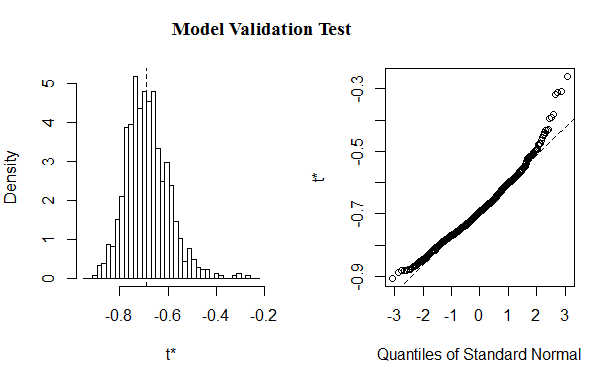
Figure 2: Shows validation test(t*) for the fitted model
The validation test (plot of the residuals), as Figure 2, shows that the simulation result obtained from bootstrap samples is suitable to estimate the model parameters.
Using our experimental data as initials samples, we applied Bootsrap statistical to sample empirical data. The simulated result is presented as Figure (1). And, the estimated model parameters (mu(µ), lamda(λ), A) are summarized as Table 1.
$$\text{Table 1: Estimated Model Parameter}$$
-------------------------------------------
mu lamda A
===========================================
Lower 0.294 16.041 1.155
Mean 0.384 16.456 1.187
Upper 0.474 16.871 1.220
Std 0.046 0.212 0.016
-------------------------------------------
Integral value: 16.622
-------------------------------------------
From the result on table $1$, the $lower$ and $upper$ values are the estimated confidence intervals of the corresponding parameters. $Std$-standard deviation) and $mean$-average values are obtained from the bootstrap samples. These results are used in the proposed three growth models. After the approprate models are selected (in our case, the three proposed models are equally approprate), the parameters are taken as an intial values for the interaction model; which is used to study the growth of S.mutant in the presence of other species(see competition model).
$$\text{Table 2: Analysis of Regression Model}$$
============================================================== Estimated parametrs: | Statistical Tests: Intercept (alpha)=7.6074 | R-squared= 0.9135 OD (beta)=-2.0365 | p-value=6.228e-14 -------------------------------------------------------------- F-statistic=254.6 ===============================================================
Using the table 2 result, the pH model can be estimated as: $$pH=7.61 - 2.04 OD$$ There is a strong linear correlation between pH and OD (R=0.9135, F=254.5, P.value $<$0.05). The coefficient value indicates that for every additional unit in OD we can expect pH to decrease by an average of 2.04. For examples: if the OD=1, pH is expected to be (pH=7.61 - 2.04(1))=5.57. The red fitted line graphically shows the same information.
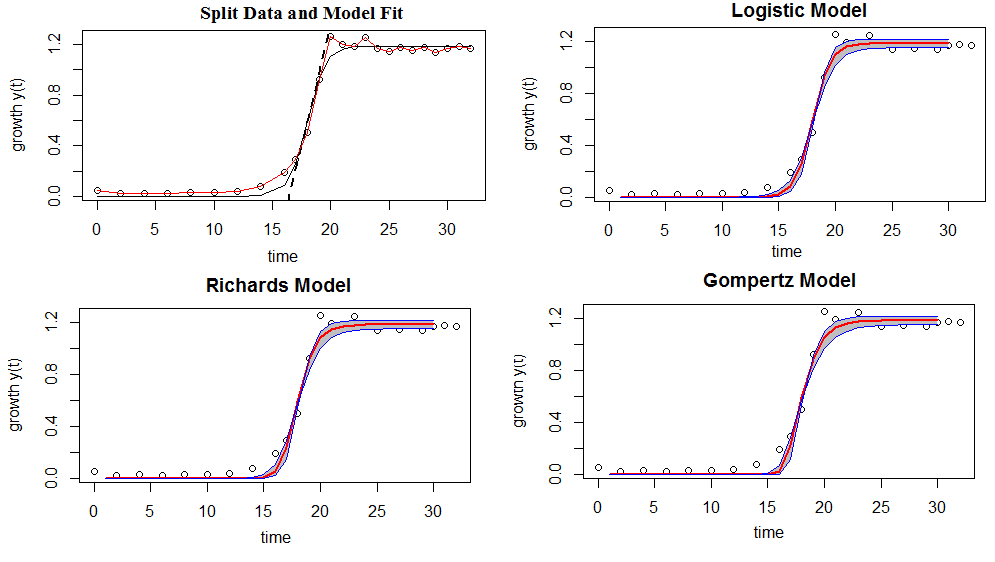
Figure 3: Shows model comparison-the shaded regions indicate a 95% confiden interval of the model fits (bold lines)
If we move left or right along the x-axis by an amount that represents a one unit change in OD, the fitted line falls by 2.04 units. If the fitted line was flat (a slope coefficient of zero), the expected value for pH would not change no matter how far up and down the line you go. So, the low p-value ($\leq$ 0.05) suggests that the slope is not zero, which in turn suggests that changes in the predictor variable(OD) are associated with changes in the response variable (pH). R-squared is a statistical measure of how close the data are to the fitted regression line. It is also known as the coefficient of determination. It is the percentage of the response variable variation that is explained by a linear model. In this case, R-square =0.9135 shows that 91.35% of the pH variation is explained by OD, and the remain 8.65 can be explained by other factors that are not considered in the regression model.

Figure 4: ph vs OD with 95% confident interval

Figure 5: Model Fit

Figure 6: Validation Test
 "
"




















