|
|
| Line 2: |
Line 2: |
| | <html> | | <html> |
| | <div id="main-content"> | | <div id="main-content"> |
| - | <h>Control-Sensor...</h> | + | <h>Control-Target</h> |
| | + | <h1>Here’s the Gist…</h1> |
| | <div class='abstract'> | | <div class='abstract'> |
| | <ul> | | <ul> |
| - | <li>We incorporate our circuit into Streptococcus phage M102, using the lysis gene of phage to kill S.mutans.</li> | + | <li>We incorporate our circuit into Streptococcus phage M102. The phage-infected Streptococcus mutans emit quorum-sensing signal will pass the message down to our probiotics terminus then trigger the S. mutans killing module of it.</li> |
| - | <li>In order to control the number of Streptococcus mutans, we use constitutive promoter to sense S.mutans by using a quorum sensing chemical compound which will be released in every competence S. mutans .</li>
| + | |
| - | <li>To control the number of S.mutans more accurately, we design the threshold composed of different terminators.</li>
| + | |
| - | <li>When the number of S.mutans reach the threshold, the lysis gene of Streptococcus phage will be triggered, and killing the S.mutans.</li>
| + | |
| | </ul> | | </ul> |
| | </div> | | </div> |
| | <div class='cont-panel'> | | <div class='cont-panel'> |
| - | <div href='#1c-1'><p>purpose</p></div> | + | <div href='#1c-1'><p>Get started.</p></div> |
| - | <div href='#1c-2'><p>background</p></div> | + | <div href='#1c-2'><p>How to do it?</p></div> |
| - | <div href='#1c-3'><p>design</p></div> | + | <div href='#1c-3'><p>Test it!</p></div> |
| - | <div href='#1c-4'><p style="line-height: 25px;">Functional Measurement</p></div> | + | <div href='#1c-4'><p>Our result~</p></div> |
| - | <div href='#1c-5'><p>result</p></div> | + | <div href='#1c-5'><p>Reference</p></div> |
| | <div style="display:inline-block; width: 640px; height: 0.1px; border: none; margin: 0px; visibility: hidden;"></div> | | <div style="display:inline-block; width: 640px; height: 0.1px; border: none; margin: 0px; visibility: hidden;"></div> |
| | </div> | | </div> |
| - | <div class='article'>
| + | <div class='article'> |
| - | <h1 id='1c-1'>Purpose</h1> | + | <h1 id='1c-1'>Before we get started:</h1> |
| - | <p>We are going to use the “control part” to control the number of Streptococcus mutans (S. mutans). When the number of S. mutans exceeds the threshold that causes cavities, the circuit will be activated, thus killing the excess S.mutans.</p> | + | <p>The "control part" is going to control the number of S. mutans. When the number of S. mutans exceeds the threshold that causes cavities, the circuit will be activated, thus killing the excess S. mutans.</p> |
| - | <h1 id='1c-2'>Background</h1>
| + | <p>In order to find and confirm the threshold that causes cavities, we designed <b>different kinds and numbers of terminators</b> (each with different leakage rates) to create the threshold. Moreover, we combined both wet lab results and modelling to decide which design is more suitable.</p> |
| - | <p>It is said that the number of S. mutans is highly related to the outbreak of cavities. Therefore, controlling the number of S. mutans has been our first priority. To reach our purpose, we need to detect the number of S. mutans, determine the threshold that causes cavities, and finally, kill the S. mutans. These three main designs of our first C, “control”, will be explained in the following paragraphs.</p> | + | <p>Over the years, it has been found that the <b>competence stimulating peptide, CSP</b>, a quorum sensing chemical, is released in every competence S. mutans. Thus, we can detect the number of S. mutans by marking the amount of CSP.</p> |
| - | <p>First of all, we need to detect the number of S. mutans. Over the years, it has been found that the <b>competence stimulating peptide, CSP</b>, a quorum sensing chemical, will be released in every competence S. mutans. Thus, we can detect the number of S.mutans by marking the amount of CSP.</p> | + | <p>In S. mutans the CSP bind to the membrane receptor, "comD", thereby phosphorylating the response regulator, "comE". The phosphorylated comE will activate numerous vital biological mechanisms in Streptococcus mutans such as biofilm formation and the release of mutacin, an antibiotic peptide. Compared with all the other mechanisms involved with CSP, it is found that the promoter of gene "nlmC"( non-lantibiotic mutacin C) in S. mutans has the best performance under the stimulation of CSP.</p> |
| - | <p>In S.mutans, the CSP will bind to the membrane receptor, “comD”, thereby phosphorylating the response regulator, “comE”. The phosphorylated comE will activate numerous vital biological mechanisms in Streptococcus mutans such as biofilm formation and mutacin release. Compared with all the other mechanisms involved with CSP, it is found that the promoter of gene “nlmC”( non-lantibiotic mutacin C) in S.mutans has the best performance under the stimulation of CSP. As a result, we have decided to use this promoter in our design.</p> | + | <img src='/wiki/images/d/dd/NYMU14_target-sensor.gif' style='width:500px;'> |
| - | <p>Secondly, we need to find and confirm the threshold that causes cavities. In this part, we will use different kinds and numbers of terminators (each with different leakage rates) to create the threshold. Moreover, we will combine both wet lab results and modelling to decide which design is more suitable.</p> | + | <h1 id='1c-2'>So how did we do it?</h1> |
| - | <p>Lastly, we need to kill the S. mutans when the amount of S. mutans exceeds the threshold. To fulfill our goal, we decided to incorporate our circuit into the Streptococcus phage M102, which is highly specific to S. mutans, in order to precisely control the lysis genes in phage M102.</p>
| + | |
| - | <h1 id='1c-3'>Design</h1> | + | <h1 id='1c-3'>Putting it to the test!</h1> |
| - | <p>!figure not yet!</p>
| + | |
| - | <p>In our circuit, we incorporate nlmC promoter and different combination of terminators. When the number of S. mutans exceed the threshold, the first “C” , control mechanism will activate the lysis genes of phage M102. Thus, control the number of S. mutans.</p>
| + | <h1 id='1c-4'>Our result</h1> |
| - | <h1 id='1c-4'>Functional Measurement</h1> | + | |
| - | <p>In order to test the function of the “control” part of this project, we incorporate two stages of functional tests.</p>
| + | <h1 id='1c-5'>Reference</h1> |
| - | <h3>The first stage</h3>
| + | <ol> |
| - | <p>We will construct two circuits. One is served as a positive control, constructed with a constitutive promoter and the GFP gene from the iGEM kit, E0040. The other is served as an experimental group, constructed with a nlmC promoter and E0040. When doing the functional measurement, we will apply different amount of synthetic CSP in different time lapse into our experimental group after the circuit being transformed into S. mutans.</p>
| + | <li>http://www.expertsmind.com/topic/microbiology/bacterial-cell-wall-92313.aspx</li> |
| - | <p>In the first stage of functional test, it is hoped to see the high amount of green fluorescent in the positive control, while different amount of green fluorescent being observed in our experimental group.</p>
| + | </ol> |
| - | <h3>The second stage</h3>
| + | </div> <!-- article --> |
| - | <p>We will test the circuit constructed with nlmC promoter and different kinds of terminator, also using E0040 to measure the threshold we create with terminators. In this stage, we will test three different terminators, B0012, B0015, and one derived from S.mutans in different combination, namely, we will construct nine circuits:</p>
| + | |
| - | <ul type='circle'> | + | |
| - | <li>nlmC promoter + B0012 + E0040</li>
| + | |
| - | <li>nlmC promoter + B0015 + E0040</li>
| + | |
| - | <li>nlmC promoter + terminator from S.mutans + E0040</li>
| + | |
| - | <li>nlmC promoter + B0012 + B0012 + E0040</li>
| + | |
| - | <li>nlmC promoter + B0012 + B0015 + E0040</li>
| + | |
| - | <li>nlmC promoter + B0012 + terminator from S.mutans + E0040</li>
| + | |
| - | <li>nlmC promoter + B0015 + B0015 + E0040</li>
| + | |
| - | <li>nlmC promoter + B0015 + terminator from S.mutans + E0040</li>
| + | |
| - | <li>nlmC promoter + terminator from S.mutans+ terminator from S.mutans +E0040</li>
| + | |
| - | </ul> | + | |
| - | <p>In the second stage of our functional test, we are looking forward to see different amount of green fluorescent after applying different amount of synthetic CSP in different time lapse into these nine circuits.</p>
| + | |
| - | <p>After the second stage of the function test, we will hand over our data to our modeling group, to see which threshold suit best in killing S.mutans.</p>
| + | |
| - | <h1 id='1c-5'>Result</h1> | + | |
| - | <h1>Reference</h1>
| + | |
| - | <ol>
| + | |
| - | <li>Kreth, J., Hung, D. C. I., Merritt, J., Perry, J., Zhu, L., Goodman, S. D., Cvitkovitch, D. G., ... Qi, F. (January 01, 2007). The response regulator ComE in Streptococcus mutans functions both as a transcription activator of mutacin production and repressor of CSP biosynthesis. Microbiology Reading-, 153, 1799-1807.</li>
| + | |
| - | <li>Hung, D. C. I., Downey, J. S., Ayala, E. A., Kreth, J., Mair, R., Senadheera, D. B., Qi, F., ... Goodman, S. D. (June 28, 2011). Characterization of DNA Binding Sites of the ComE Response Regulator from Streptococcus mutans. Journal of Bacteriology, 193, 14, 3642-3652.</li>
| + | |
| - | <li>Liu, T., Xue, S., Cai, W., Liu, X., Liu, X., Zheng, R., Luo, H., ... Qi, W. (March 22, 2014). ComCED signal loop precisely regulates nlmC expression in Streptococcus mutans. Annals of Microbiology, 64, 1, 31-38.</li> | + | |
| - | <li>van der Ploeg, J R. (2007). Genome sequence of Streptococcus mutans bacteriophage M102. Wiley-Blackwell.</li>
| + | |
| - | </ol>
| + | |
| - | </div> <!-- article -->
| + | |
| | </div> | | </div> |
| | </html> | | </html> |
| | {{:Team:NYMU-Taipei/NYMU14_Footer}} | | {{:Team:NYMU-Taipei/NYMU14_Footer}} |
Control-Target
Here’s the Gist…
- We incorporate our circuit into Streptococcus phage M102. The phage-infected Streptococcus mutans emit quorum-sensing signal will pass the message down to our probiotics terminus then trigger the S. mutans killing module of it.
Before we get started:
The "control part" is going to control the number of S. mutans. When the number of S. mutans exceeds the threshold that causes cavities, the circuit will be activated, thus killing the excess S. mutans.
In order to find and confirm the threshold that causes cavities, we designed different kinds and numbers of terminators (each with different leakage rates) to create the threshold. Moreover, we combined both wet lab results and modelling to decide which design is more suitable.
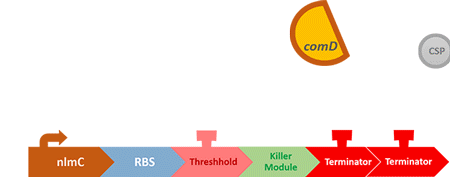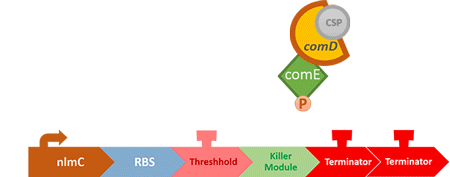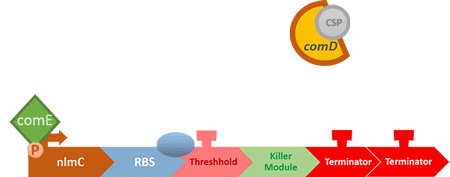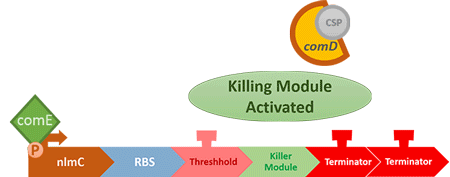
Over the years, it has been found that the competence stimulating peptide, CSP, a quorum sensing chemical, is released in every competence S. mutans. Thus, we can detect the number of S. mutans by marking the amount of CSP.
In S. mutans the CSP bind to the membrane receptor, "comD", thereby phosphorylating the response regulator, "comE". The phosphorylated comE will activate numerous vital biological mechanisms in Streptococcus mutans such as biofilm formation and the release of mutacin, an antibiotic peptide. Compared with all the other mechanisms involved with CSP, it is found that the promoter of gene "nlmC"( non-lantibiotic mutacin C) in S. mutans has the best performance under the stimulation of CSP.
So how did we do it?
Putting it to the test!
Our result
Reference
- http://www.expertsmind.com/topic/microbiology/bacterial-cell-wall-92313.aspx

 "
"




















