|
|
| (23 intermediate revisions not shown) |
| Line 2: |
Line 2: |
| | <html> | | <html> |
| | <div id="main-content"> | | <div id="main-content"> |
| - | <h>TARGET</h> | + | <h>Control-<span style='color:rgb(249, 103, 236);'>Target</span></h> |
| - | | + | <h1>Here’s the Gist…</h1> |
| - | <h1>Here’s the Gist..</h1> | + | <div class='abstract'> |
| - | <div class="abstract"> | + | |
| | <ul> | | <ul> |
| - | <li> Utilize the <i>Streptococcus phage</i> M102 to infect <i>S. mutans</i> and act as a sensor.</li>
| + | <li>Utilize the <i>Streptococcus</i> phage M102 to infect <i>S. mutans</i> and act as a sensor.</li> |
| - | <li> When <i>S. mutans</i> population exceeds a threshold, M102 sends out a quorum-sensing signal to our probiotic.</li> | + | <li>When <i>S. mutans</i> population exceeds a threshold, <i>M102</i> sends out a quorum-sensing signal to our probiotic. |
| - | <li> The signal turns on the probiotic's killing module and bring down <i>S. mutans</i> population.</li> | + | <li>This signal then turns on the probiotic’s killing module and brings down the <i>S. mutans</i> population. |
| | + | </li> |
| | </ul> | | </ul> |
| | </div> | | </div> |
| Line 18: |
Line 18: |
| | <div href='#1c-4'><p>Our result~</p></div> | | <div href='#1c-4'><p>Our result~</p></div> |
| | <div href='#1c-5'><p>Reference</p></div> | | <div href='#1c-5'><p>Reference</p></div> |
| - | <div style="display:inline-block; width: 640px; height: 0.1px; border: none; margin: 0px"></div> | + | <div style="display:inline-block; width: 640px; height: 0.1px; border: none; margin: 0px; visibility: hidden;"></div> |
| | </div> | | </div> |
| - | <div class='article indent'>
| + | <div class='article indent'> |
| - | <h1 id='1c-1'>Before we get started:</h1> | + | <h1 id='1c-1'>Before we get started:</h1> |
| - | <p>Control is so-called because it is used to control the number of <i>S. mutans</i>, instead of killing it completely off, as indicated by [<a href='/Team:NYMU-Taipei/modeling/m2'>Modeling: Competition</a>]. When the number of <i>S. mutans</i> exceeds the threshold that causes cavities, the circuit will be activated, thus killing the excess <i>S. mutans</i>.</p> | + | <p>"Control" is so-called because it is used to control the number of <i>S. mutans</i>, instead of killing it completely off, which wouldn't help the matter as indicated in [<a href='/Team:NYMU-Taipei/modeling/m2'>Modeling: Competition</a>]. When the number of <i>S. mutans</i> exceeds the threshold that causes cavities, the circuit will be activated, thus killing the excess <i>S. mutans</i>.</p> |
| - | | + | <p>In order to find and confirm the threshold that causes cavities, we designed <b>different kinds and numbers of terminators</b> (each with different leakage rates) to create the threshold. Moreover, we combined both wet lab results and modelling to decide which design is more suitable.</p> |
| - | <p>In order to find and confirm the threshold that causes cavities, we designed <b>different kinds and numbers of terminators</b> (each with different leakage rates) to create the threshold. Moreover, we combined both wet lab results and modelling to decide which design is more suitable. </p> | + | <p>Over the years, it has been found that the <b>competence stimulating peptide, CSP</b>, a quorum sensing chemical, is released in every competence <i>S. mutan</i>s. Thus, we can detect the number of <i>S. mutans</i> by marking the amount of CSP.</p> |
| - | | + | <img src='/wiki/images/d/dd/NYMU14_target-sensor.gif'> |
| - | <p>Over the years, it has been found that the <b>competence stimulating peptide, CSP</b>, a quorum sensing chemical, is released in every competence <i>S. mutans</i>. Thus, we can detect the number of <i>S. mutans</i> by marking the amount of CSP. </p> | + | <p>In <i>S. mutans</i> the CSP bind to the membrane receptor, "comD", thereby phosphorylating the response regulator, "comE". The phosphorylated comE will activate numerous vital biological mechanisms in Streptococcus mutans such as biofilm formation and the release of mutacin, an antibiotic peptide. Compared with all the other mechanisms involved with CSP, it is found that the promoter of gene "nlmC"( non-lantibiotic mutacin C) in S. mutans has the best performance under the stimulation of CSP.</p> |
| - | | + | <p>At this point, the two main organisms we adapted for our project are our probiotics and <i>M102</i> bacteriophage. We used one of the most commonly seen <b>quorum sensing chemicals, AHL</b>, N-Acyl-homoserine-lactone, <b>as our indicator between phage and E.coli</b>. Due to the idea of phage threshold control of the number of <i>S. mutans</i> in prevention of the second-dominant strain in the oral cavity, we envision a possibility to trigger another killing system to reach synergistic effect on diminishing <i>S. mutans</i>. If the threshold terminators were passed, the downstream signaling pathway would be switched on and contact with the engineered probiotics.</p> |
| - | <p>In <i>S. mutans</i> the CSP bind to the membrane receptor, "comD", thereby phosphorylating the response regulator, "comE". The phosphorylated comE will activate numerous vital biological mechanisms in Streptococcus mutans such as biofilm formation and the release of mutacin, an antibiotic peptide. Compared with all the other mechanisms involved with CSP, it is found that the promoter of gene "nlmC"( non-lantibiotic mutacin C) in <i>S. mutans</i> has the best performance under the stimulation of CSP. </p> | + | <p>When the amount of the <i>S. mutans</i> is so large that the phages couldn't afford the loading, CSP would pass the threshold value and turn on the <b>luxR-luxI system</b>. LuxI acts as a gene which would generate AHL-synthase that yielding the quorum-sensing signal AHL right after the threshold level of <i>S. mutans</i> is exceeded. LuxR is a protein which forms a complex with AHL, and then turns into an activated form which will afterwards bind with the promoter pLuxR and trigger it. |
| - | [放gift]
| + | |
| - | | + | |
| - | <p>At this point, the two main organisms we adapted for our project are E. coli and M102 bacteriophage. We used one of the most commonly seen <b>quorum sensing chemicals, AHL</b>, N-Acyl-homoserine-lactone, <b>as our indicator between phage and <i>E.coli</i>. </b> Due to the idea of phage threshold control of the number of <i>S. mutans</i> in prevention of the second-dominant strain in the oral cavity, we envision a possibility to trigger another killing system to reach synergistic effect on diminishing <i>S. mutans</i>. If the threshold terminators were passed, the downstream signaling pathway would be switched on and contact with the engineered <i>E.coli</i>.</p> | + | |
| - | <p>When the amount of the <i>S. mutans</i> is so large that the phages couldn't afford the loading, CSP would pass the threshold value and turn on the <b>luxR-luxI system. </b> LuxI acts as a gene which would generate AHL-synthase that yielding the quorum-sensing signal AHL right after the threshold level of <i>S. mutans</i> is exceeded. LuxR is a protein which forms a complex with AHL, and then turns into an activated form which will afterwards bind with the promoter pLuxR and trigger it. | + | |
| | The signal sequence and endolysin downstream would be secreted and eliminate the excess <i>S. mutans</i>.</p> | | The signal sequence and endolysin downstream would be secreted and eliminate the excess <i>S. mutans</i>.</p> |
| | + | <div style='float:right'><img src='/wiki/images/2/2c/NYMU14_target_cellwall.png' style='width: 450px;padding-left: 20px;'><p style='width: 450px;font-size: 14px;'>Figure 1. Gram positive bacteria have only one layer of cell wall outside of cell membrane, composing of peptidoglycan, while the cell wall of gram negative bacteria has additional protection outside peptidoglycan cell wall.[1]</p></div> |
| | + | <p>Endolysin is an enzyme expressed by phage-infected bacteria. The C-terminus bind to the host cell wall, while the N-terminus is enzymatic domain. Working with holin, which lyses cell membrane, endolysin could break the cell wall of bacteria. Usually serve as peptidase, endolysin tend to work on gram positive species, which have layers of peptidoglycan cell wall. Though originally intended to be utilized inside the cell wall, endolysin can also be applied outside cell according to previous research. Furthermore, it is even less harmful to gram negative bacteria this way because of the lipid layer of those bacteria composes cell wall.</p> |
| | + | |
| | + | <h1 id='1c-2' style='float:left;'>So how did we do it?</h1> |
| | + | <img src='/wiki/images/f/fa/NYMU14_target_sensor_ter.png' style='width:500px;'> |
| | + | <p>In our circuit, we incorporate nlmC promoter and <b>different combination of terminators to create the threshold</b>.</p> |
| | + | <p>When the amount of <i>S. mutans</i> is so large that the phages couldn't afford the loading, CSP would pass the threshold value and turn on the <b>luxR-luxI system</b>. LuxI in <i>S. mutans</i> will be triggered by nlmC and activate AHL which will combine with LuxR produced by probiotics continuously. When LuxR combine with AHL to form AHL-LuxR complex, it will induce pLuxR promoter and trigger the Killer Module.</p> |
| | + | <img src='/wiki/images/3/31/NYMU14_targetall.png' style='width:760px;'> |
| | + | <h3>Killer Module</h3> |
| | + | <img src='/wiki/images/a/a7/NYMU_killing_module.png' style='width:580px;'> |
| | + | <p>YebF is a protein that is naturally secreted by <i>E.coli</i>. It would transport proteins attached to it to the periplasm (space between cell membrane and cell wall), and later secrete it through porin on the cell wall. We chose YebF for its two advantages: one, it can be secreted in experimental strains which usually do not secrete proteins, along with other diverse passenger characters. Two, YebF can prevent endolysin from toxicating <i>E. coli</i>.</p> |
| | | | |
| - | <p>Endolysin is an enzyme expressed by phage-infected bacteria. The C-terminus bind to the host cell wall, while the N-terminus is enzymatic domain. Working with holin, which lyses cell membrane, endolysin could break the cell wall of bacteria. Usually serve as peptidase, endolysin tend to work on gram positive species, who have layers of peptidoglycan cell wall. Though originally intended to be utilized inside the cell wall, endolysin can also be applied outside cell according to previous research. Furthermore, it is even less harmful to gram negative bacteria this way because of the lipid layer of those bacteria composes cell wall.</p>
| + | <h1 id='1c-3'>Putting it to the test!</h1> |
| - | <div><img src="/wiki/images/a/a6/NYMU14_Target_FIG1.jpg" style="width: 480px;"><p style="font-size: 14px;display: block; margin: 0 auto; text-align: center;">Figure 1. </p></div>
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | <p>Gram positive bacteria has only one layer of cell wall outside of cell membrane, composing of peptidoglycan.</p>
| + | |
| - | <p>While the cell wall of gram negative bacteria has additional protection outside peptidoglycan cell wall.[1]</p>
| + | |
| - | | + | |
| - | <h1 id='1c-2'>So how did we do it? </h1>
| + | |
| - |
| + | |
| - | <img src="/wiki/images/6/6c/NYMU14_Target_T2-1.png">
| + | |
| - | <img src="/wiki/images/3/3e/NYMU14_Target_T2-2.png">
| + | |
| - |
| + | |
| - | <p>In our circuit, we incorporate nlmC promoter and <b>different combination of terminators to create the threshold. </b></p>
| + | |
| - | | + | |
| - | <p>When the amount of <i>S. mutans</i> is so large that the phages couldn't afford the loading, CSP would pass the threshold value and turn on the <b>luxR-luxI system</b>.</p>
| + | |
| - | <p>LuxI in <i>S. mutans</i> will be triggered by nlmC and activate AHL which will combine with LuxR produced by E. coli continuously. When LuxR combine with AHL to form AHL-LuxR complex, it will induce pLuxR promoter and trigger the Killer Module. </p>
| + | |
| - | <img src="/wiki/images/1/10/NYMU14_Target_FIG2.png">
| + | |
| - |
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | <h3>Killer Module</h3>
| + | |
| - | <img src="/wiki/images/8/8d/NYMU14_Target_T6-1.png">
| + | |
| - |
| + | |
| - | <h3>YebF</h3>
| + | |
| - | <p>YebF is a protein that is naturally secreted by <i>E.coli</i>. It would transport proteins attached to it to the periplasm (space between cell membrane and cell wall), and later secrete it through porin on the cell wall. We chose YebF for its two advantages: one, it can be secreted in experimental strains which usually do not secrete proteins, along with other diverse passenger characters. Two, YebF can prevent endolysin from toxicating <i>E.coli</i>.</p>
| + | |
| - | | + | |
| - | | + | |
| - | <h1 id='1c-3'>Putting it to the test! </h1> | + | |
| - | ((Sensor))
| + | |
| | <h3>Stage one: How well does nlmC function?</h3> | | <h3>Stage one: How well does nlmC function?</h3> |
| - | <p>In order to test if nlmC functions as we expected it to, we constructed two circuits. The first is constructed with a constitutive promoter and GFP, and will serve as our control: </p> | + | <p>In order to test if nlmC functions as we expected it to, we constructed two circuits. The first is constructed with a constitutive promoter and GFP, and will serve as our control:</p> |
| - | <img src="/wiki/images/d/da/NYMU14_Target_T1.png"> | + | <img src='/wiki/images/d/da/NYMU14_Target_T1.png' style='width: 300px;'> |
| - |
| + | <p>The other is constructed instead with nlmC and GFP, and will serve as the testing circuit for comparison between the efficacy of a constitutive promoter and the nlmC promoter:</p> |
| - | <p>The other is constructed instead with nlmC and GFP, and will serve as the testing circuit for comparison between the efficacy of a constitutive promoter and the nlmC promoter: </p> | + | <img src='/wiki/images/b/b5/NYMU14_Target_T1-2.png' style='width: 300px;'> |
| - | <img src="/wiki/images/b/b5/NYMU14_Target_T1-2.png"> | + | <p>To compare these circuits, we applied different amounts of synthetic CSP in different time lapses to our experimental group. If everything works as planned, the experimental group should display fluorescence in a wide array of intensity depending on CSP concentration, while the control group displays a uniform bright green.</p> |
| - |
| + | |
| - | <p>To compare these circuits, we applied different amounts of synthetic CSP in different time lapses to our experimental group. If everything works as planned, the experimental group should display fluorescence in a wide array of intensity depending on CSP concentration, while the control group displays a uniform bright green. </p> | + | |
| | | | |
| | <h3>Stage two: Which threshold combination should we use?</h3> | | <h3>Stage two: Which threshold combination should we use?</h3> |
| - | <p>After confirming that the nlmC promoter works and responds to different levels of CSP, we need to design a test to determine which terminator combination we can best use as an effective threshold. We designed two circuits with different terminator thresholds, B0012 and double B0012, respectively, and GFP after them as a reporter. </p> | + | <p>After confirming that the nlmC promoter works and responds to different levels of CSP, we need to design a test to determine which terminator combination we can best use as an effective threshold. We designed two circuits with different terminator thresholds, B0012 and double B0012, respectively, and GFP after them as a reporter.</p> |
| - |
| + | <img src='/wiki/images/f/fa/NYMU14_target_sensor_ter.png' style='width:500px;'> |
| - | <img src="/wiki/images/3/3e/NYMU14_Target_T2-2.png"> | + | <p>The testing is similar to Stage 1, and we simply recorded the GFP production after induction with commercial CSP.</p> |
| - | <img src="/wiki/images/6/6c/NYMU14_Target_T2-1.png">
| + | <p>In the future, this protocol can be used to test many other combinations to find the "perfect" threshold for HOPE. After obtaining some experimental data, we employed extensive modeling to incorporate these numbers into other combinations of terminators, and we’ll be looking to validate these results after the Jamboree is over to further strengthen HOPE. Please see our <a href='Team:NYMU-Taipei/modeling/m3'>[Model: Threshold]</a> section for more information.</p> |
| - | | + | |
| - |
| + | |
| - | <p>The testing is similar to Stage 1, and we simply recorded the GFP production after induction with commercial CSP. </p> | + | |
| - | | + | |
| - | <p>In the future, this protocol can be used to test many other combinations to find the "perfect" threshold for HOPE. After obtaining some experimental data, we employed extensive modeling to incorporate these numbers into other combinations of terminators, and we'll be looking to validate these results after the Jamboree is over to further strengthen HOPE. Please see our [Model: Threshold] section for more information. </p> | + | |
| - | | + | |
| - | ((communication))
| + | |
| | | | |
| | <h3>Stage three: Mechanisms of the Signal Receptor in our Probiotic</h3> | | <h3>Stage three: Mechanisms of the Signal Receptor in our Probiotic</h3> |
| | <p>After all the fragments of the circuit is linked, we need a test to tell us that the fragments are well-connected, and that the mechanisms linking them has not lost its function. We created a circuit which is identical to our final circuit, except with RFP in place of the killing module:</p> | | <p>After all the fragments of the circuit is linked, we need a test to tell us that the fragments are well-connected, and that the mechanisms linking them has not lost its function. We created a circuit which is identical to our final circuit, except with RFP in place of the killing module:</p> |
| - | <img src="/wiki/images/8/8c/NYMU14_Target_T3.png"> | + | <img src='/wiki/images/8/8c/NYMU14_Target_T3.png'> |
| - | | + | |
| - |
| + | |
| | <p>We then transformed our circuit into <i>E. coli</i>, and added in commercial AHL. If the mechanisms remain robust after splicing, we should see a strong display of red from RFP.</p> | | <p>We then transformed our circuit into <i>E. coli</i>, and added in commercial AHL. If the mechanisms remain robust after splicing, we should see a strong display of red from RFP.</p> |
| - | <img src="/wiki/images/2/2a/NYMU14_Target_Construction_Check_2.png"> | + | <img src='/wiki/images/2/2a/NYMU14_Target_Construction_Check_2.png' style='width: 300px;margin-left: 40px;'> |
| - |
| + | |
| | <h3>Stage four: Mechanisms of the Signal Producer in <i>S. mutans</i></h3> | | <h3>Stage four: Mechanisms of the Signal Producer in <i>S. mutans</i></h3> |
| - | <p>One main concern with our mechanism is that AHL is a Gram-negative quorum sensing-signal… and yet we are implanting it into the Gram-positive <i>S. mutans</i> via <i>M102</i>. We need to test if the AHL production still works in <i>S. mutans</i>.</p> | + | <img src='/wiki/images/b/b2/NYMU14_Target_Test2.png' style='float: right;width: 340px;padding-left: 20px;padding-bottom: 20px;padding-top: 20px;'> |
| | + | <p>One main concern with our mechanism is that AHL is a Gram-negative quorum sensing-signal… and yet we are implanting it into the Gram-positive <i>S.mutans</i> via <i>M102</i>. We need to test if the AHL production still works in <i>S. mutans</i>.</p> |
| | + | <p>In order to confirm that whether the engineered circuit could successfully yield AHL, we adopt the purification and structural examination processes referred to related references.The LuxI gene we embedded inside our circuit coded BBa_C0061 in IGEM registry and it synthesizes N-3-(Oxohexanoyl)-L-homoserine lactone. We uses preparative-TLC to purify the chemical and conduct 1HNMR to check the structure.</p> |
| | + | <p>According to paper research, we prepared our sample through the protocol described below:</p> |
| | + | <ol> |
| | + | <li>S. mutans liquid cultured till its stationary phase</li> |
| | + | <li>Add lysozyme to 5ml cultures</li> |
| | + | <li>Sonication</li> |
| | + | <li>Centrifugation</li> |
| | + | <li>Extract the supernatant with equal volume of ethyl acetate X2</li> |
| | + | <li>Dry with anhydrous magnesium sulfate</li> |
| | + | <li>Filtering</li> |
| | + | <li>Evaporate to dry</li> |
| | + | <li>Dissolve with 50-100uL HPLC-grade ethyl acetate</li> |
| | + | <li>Run TLC</li> |
| | + | </ol> |
| | | | |
| - | <p>In this experiment, we first made a standard curve of the concentration of commercialized AHL versus absorption value. After that, we transformed the circuit containing LuxI into <i>S. mutans</i>. By extracting the supernatant of the culturing medium of the engineered <i>S. mutans</i>, and using a spectrometer to measure the concentration of AHL compared to the standard curve made previously, we can determine whether or not AHL is being expressed successfully in <i>S. mutans</i>.</p>
| |
| - | <img src="/wiki/images/2/2a/NYMU14_Target_Construction_Check_2.png">
| |
| - |
| |
| - | ((killing))
| |
| | <h3>Stage five: Does YebF work?</h3> | | <h3>Stage five: Does YebF work?</h3> |
| | <p>Before we check the functionality of the entire construct, we need to test the YebF protein for its ability to transport attached parts out of the membrane. We began by constructing two circuits:</p> | | <p>Before we check the functionality of the entire construct, we need to test the YebF protein for its ability to transport attached parts out of the membrane. We began by constructing two circuits:</p> |
| - | <img src="/wiki/images/d/d2/NYMU14_Target_T5-1.png"> | + | <img src='/wiki/images/d/d3/NYMU14_target_YebF_t.png' style='width: 550px;'> |
| - |
| + | <p>These circuits are then transformed into <i>E. coli</i>, cultivated in liquid culture, and centrifuged. Afterwards, the supernatants are analyzed for flourescence intensity fluorescence intrnsity and their OD value. If YebF works as we expected, we should see the first circuit successfully secrete RFP into the supernatant :</p> |
| - | <img src="/wiki/images/0/07/NYMU14_Target_T5-2.png">
| + | <img src='/wiki/images/a/a6/NYMU14_target_YebF_rfp.png' style='width: 220px;margin-left: 40px;'> |
| - |
| + | |
| - | <p>These circuits are then transformed into <i>E. coli</i>, cultivated in liquid culture, and centrifuged. Afterwards, the supernatants are analyzed for their OD value. If YebF works as we expected, we should see the first circuit successfully secrete RFP into the supernatant:</p> | + | |
| - | <img src="/wiki/images/6/67/NYMU14_Target_1.png" style="width: 120px; display: inline-block;"> | + | |
| - | <img src="/wiki/images/f/f9/NYMU14_attach_Result3.png" style="width: 120px;">
| + | |
| | <p>While the control should only contain RFP which is not exported:</p> | | <p>While the control should only contain RFP which is not exported:</p> |
| - | <img src="/wiki/images/4/48/NYMU14_Target_2.png" style="width: 120px; display: inline-block;"> | + | <img src='/wiki/images/8/87/NYMU14_target_YebF_norfp.png' style='width: 190px;margin-left: 40px;'> |
| - | <img src="/wiki/images/4/45/NYMU14_attach_Result2.png" style="width: 120px;">
| + | |
| | <h3>Stage six: Does Endolysin work?</h3> | | <h3>Stage six: Does Endolysin work?</h3> |
| | <p>To test the antimicrobial activity of endolysin, we also created two circuits:</p> | | <p>To test the antimicrobial activity of endolysin, we also created two circuits:</p> |
| - | <img src="/wiki/images/8/8d/NYMU14_Target_T6-1.png"> | + | <img src='/wiki/images/d/d1/NYMU14_target_endolysin_t.png' style='width: 550px;'> |
| - |
| + | |
| - | <img src="/wiki/images/e/e5/NYMU14_Target_T6-2.png">
| + | |
| | <p>The first circuit contains YebF and endolysin, which serves as the experimental circuit, and the second contains just YebF, and is the control. These are transformed into <i>E. coli</i>, and after culturing them overnight and extracting the supernatant, we spread the supernatant on plates of <i>S. mutans</i> to test for their antimicrobial activity.</p> | | <p>The first circuit contains YebF and endolysin, which serves as the experimental circuit, and the second contains just YebF, and is the control. These are transformed into <i>E. coli</i>, and after culturing them overnight and extracting the supernatant, we spread the supernatant on plates of <i>S. mutans</i> to test for their antimicrobial activity.</p> |
| | | | |
| - | <h3>Stage five: Does AHL stimulate the killing program as anticipated?</h3> | + | <h3>Stage seven: Does AHL stimulate the killing program as anticipated?</h3> |
| - | <p>In this stage, we put the final probiotic circuit into <i>E. coli</i> to see how well it performs. </p> | + | <p>In this stage, we put the final probiotic circuit into <i>E. coli</i> to see how well it performs.</p> |
| - | <img src="/wiki/images/3/39/NYMU14_Target_T7.png"> | + | <img src='/wiki/images/3/39/NYMU14_Target_T7.png' style='width: 700px;'> |
| - |
| + | <p>The two different germs lines are involved in this test, the <i>E.coli</i> constructed circuit (<b>pSB1C3 + luxR + pluxR + YebF + Endolysin</b>) and <i>S.mutans</i> constructed circuit (luxI). |
| - | <p>We then incubated <i>S. mutans</i> together with the transformed <i>E. coli</i> and added commercial AHL to the mix. If everything works as expected, then the killing module should be successfully activated, and we should see a difference in <i>S. mutans</i> population. </p> | + | |
| - | <img src="/wiki/images/b/ba/NYMU14_Target_Test5.png">
| + | |
| - |
| + | |
| | | | |
| - | <h3>Stage six: Finally, let's check <i>Everything</i>!</h3>
| + | The culturing of constructed <i>E.coli</i> with the supernatant of constructed <i>S.mutans</i> (containing AHL) supposed to be secretion of endolysin. Then we cultured the wild type <i>S.mutans</i> with the supernatant of previous culture.</p> |
| - | <p>In this last stage we put all the constructed circuits intended for our probiotic and <i>M102</i> into <i>E. coli</i> and <i>S. mutans</i>, respectively. Then we co-cultured these two transformed strains together to test our full circuit. </p>
| + | <img src='/wiki/images/b/ba/NYMU14_Target_Test5.png' style='width: 500px;margin-left: 40px;'> |
| | | | |
| - | <h1 id='1c-4'>Our result</h1> | + | <h3>Stage eight: Finally, let’s check <i>Everything</i>!</h3> |
| - | <h1 id='1c-5'>Reference</h1> | + | <p>In this last stage we put all the constructed circuits intended for our probiotic and <i>M102</i> into <i>E. coli</i> and <i>S. mutans</i>, respectively. Then we co-cultured these two transformed strains together to test our full circuit.</p> |
| | + | |
| | + | <h1 id='1c-4'>Our results</h1> |
| | + | <h3>nlmC promoter</h3> |
| | + | <ul> |
| | + | <li>Constitutive promoter+ RBS+ GFP</li> |
| | + | <li>nlmC promoter+ RBS+ GFP</li> |
| | + | <li>nlmC promoter+ RBS+B0012+GFP</li> |
| | + | <li>nlmC promoter+ RBS+ B0012+B0012+GFP</li> |
| | + | </ul> |
| | + | <p>Study done by Liu et al. shown that nlmC promoter can be induced by CSP[1]</p> |
| | + | <img src='/wiki/images/d/d9/NYMU14_target_paperres.JPG' style='width: 500px;float: left;padding-right: 10px;'> |
| | + | <p>First of all, “constitutive promoter+ RBS+ GFP” and “nlmC promoter+ RBS+ GFP” will be tested by observing the green fluorescence in the gene-modified Streptococcus mutans. “Constitutive promoter+ RBS+ GFP” is believed to create green fluorescence constitutively without addition stimulation. On the other hand, “nlmC promoter+ RBS +GFP” is believed to create green fluorescence under the stimulation of CSP. In this step of function test, we are hoping to test the function of nlmC promoter.</p> |
| | + | <p>Secondly, “nlmC promoter+ RBS+ GFP”, “nlmC promoter+ RBS +B0012+ GFP”, and “nlmC promoter +RBS+ B0012+ B0012+GFP” will be tested to see which create the most suitable threshold for preventing cavities. It is believed that the more terminators we added to our circuit, the higher threshold will be made. Under the addition of CSP, the nlmC promoter will be induced, thereby create the green fluorescence which we can observe in the gene-modified Streptococcus mutans.</p> |
| | + | |
| | + | <h3>Testing YebF</h3> |
| | + | <p>From left to right: J23100+yebF+RFP+B0015, J23100, Competent cell</p> |
| | + | <img src='/wiki/images/a/a8/NYMU14_target_yefb1.JPG' style='width: 550px;'> |
| | + | <p>Partially enlargement:</p> |
| | + | <img src='/wiki/images/a/a1/NYMU14_target_yefb2.JPG' style='width: 550px;'> |
| | + | <p>We can see that although there is small difference, there are mostly more fluorescence in our circuit, showing that there might be little RFP secreted. But the effect is not very clear, resulting in small difference in fluorescent intensity. |
| | + | Also, the comparison between our testing circuit with other control shows that there might exist some technical problems, for sometimes LB or competent cell result in more intensity in RFP wavelength range.</p> |
| | + | <p>At the same time, measuring OD600 to make comparison : </p> |
| | + | <img src='/wiki/images/5/5a/NYMU14_target_yefb3.JPG' style='width: 550px;'> |
| | + | <p>Overall our testing circuit showed the relationship between growth and its secretion as shown below:</p> |
| | + | <img src='/wiki/images/f/fc/NYMU14_target_yefb4.JPG' style='width: 550px;'> |
| | + | <p>As seen in the graph, the secretion of YebF and RFP basically soared with the growth of the E.coli, and come to a dynamic balance at the as it entered log phase.</p> |
| | + | |
| | + | <h3>Testing Endolysin</h3> |
| | + | <p>By doing disc plate assay of endolysin together with several other controls, we can see that there is a clear space besides the disc of our different concentration of our endolysin supernatant.</p> |
| | + | <img src='/wiki/images/c/cd/NYMU14_target_endolysin_res1.png' style='width: 370px;float: left;padding: 10px;'> |
| | + | <p>Here are our samples:</p> |
| | + | <ul> |
| | + | <li>Ampicillin : 100ng/ul</li> |
| | + | <li>BHI broth</li> |
| | + | <li>J+Y+B: J23100+yebF+B0015 overnight culture supernatant</li> |
| | + | <li>(OD600=2.647)(2.0 rcf, 7min) </li> |
| | + | <li>J23100+yebF+endolysin+B0015:overnight culture supernatant </li> |
| | + | <li>(OD600=2.900)(2.0 rcf, 7min)</li> |
| | + | </ul> |
| | + | <p>Western blot filter paper:</p> |
| | + | <ul> |
| | + | <li>Positive control: Ampicillin (100ng/ul),50ul</li> |
| | + | <li>Negative control: BHI, 50ml /J23100+yebF+B0015, 50ul</li> |
| | + | <li>Testing: |
| | + | <ul> |
| | + | <li>J23100+yebF+endolysin+B0015, 50ul</li> |
| | + | <li>J23100+yebF+endolysin+B0015, 100ul</li> |
| | + | <li>J23100+yebF+endolysin+B0015, 150ul</li> |
| | + | </ul> |
| | + | </li> |
| | + | </ul> |
| | + | <p>As shown in the figure, ampicillin showed a significant inhibition zone as a positive control. Three positive control all show there ability to kill <i>Streptococcus mutans</i> by the clear zone around the disc. The BHI negative control showed no killing ability as expected.</p> |
| | + | <p>However, there are also little <i>Streptococcus mutans</i> growing near J23100+yebF+B0015 culture supernatant. By observing the pattern on the plate, we speculated that it might be the result of the diffusion of Ampicillin beside it.</p> |
| | + | |
| | + | <h3>AHL induced <i>S. mutans</i> death</h3> |
| | + | <img src='/wiki/images/c/cb/NYMU14_target_ahl.JPG' style='width: 150px;'> |
| | + | <p>The supernatant purified from the <i>S. mutans</i> (with constructed circuit: S-IT/PVA838) contains AHL that could possible trigger the killer circuit in <i>E. coli </i>and show a decreasing growth of wild type <i>S. mutans.</i></p> |
| | + | <p>Our constructed circuit (Termed E-C1) including <i>E. coli</i> promoter (Truncated BBa_K523013) region, luxR(BBa_C0062) coding sequence and pLuxR(BBa_R0062) promoter region would be 3117 bp in length. The result of constructed circuit ligation is shown below.</p> |
| | + | |
| | + | |
| | + | |
| | + | </div> <!-- article --> |
| | + | <h1 id='1c-5'>Reference</h1> |
| | <ol> | | <ol> |
| - | <li>http://www.expertsmind.com/topic/microbiology/bacterial-cell-wall-92313.aspx</li> | + | <li>Liu, T., et al., ComCED signal loop precisely regulates nlmC expression in Streptococcus mutans. Annals of Microbiology, 2014. 64(1): p. 31-38.</li> |
| | + | <li>(2006)"Extracellular accumulation of recombinant proteins fused to the carrier protein YebF in Escherichia coli"</li> |
| | + | <li>(2007)"ACES™ Signal Sequence and YebF Expression Systems Technical Brief"</li> |
| | + | <li>(2012)"A Protein Export Pathway Involving Escherichia coli Porins"</li> |
| | + | <li>(2006)patent" PROTEIN PRODUCTION METHOD UTILIZING YEBF "</li> |
| | + | <li>(2007)"Genome sequence of Streptococcus mutans bacteriophage M102"</li> |
| | + | <li>(2012)''Biology and Genome Sequence of Streptococcus mutans Phage M102AD''</li> |
| | </ol> | | </ol> |
| - |
| |
| - | </div> <!-- article -->
| |
| | </div> | | </div> |
| | </html> | | </html> |
| | {{:Team:NYMU-Taipei/NYMU14_Footer}} | | {{:Team:NYMU-Taipei/NYMU14_Footer}} |
Control-Target
Here’s the Gist…
- Utilize the Streptococcus phage M102 to infect S. mutans and act as a sensor.
- When S. mutans population exceeds a threshold, M102 sends out a quorum-sensing signal to our probiotic.
- This signal then turns on the probiotic’s killing module and brings down the S. mutans population.
Before we get started:
"Control" is so-called because it is used to control the number of S. mutans, instead of killing it completely off, which wouldn't help the matter as indicated in [Modeling: Competition]. When the number of S. mutans exceeds the threshold that causes cavities, the circuit will be activated, thus killing the excess S. mutans.
In order to find and confirm the threshold that causes cavities, we designed different kinds and numbers of terminators (each with different leakage rates) to create the threshold. Moreover, we combined both wet lab results and modelling to decide which design is more suitable.
Over the years, it has been found that the competence stimulating peptide, CSP, a quorum sensing chemical, is released in every competence S. mutans. Thus, we can detect the number of S. mutans by marking the amount of CSP.
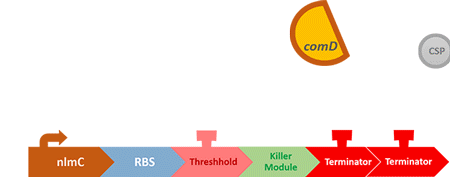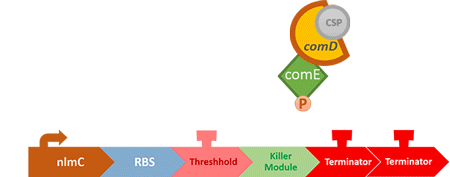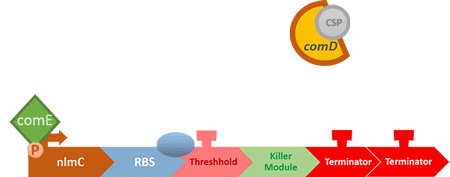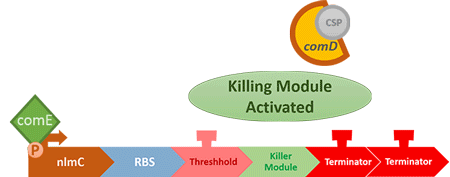
In S. mutans the CSP bind to the membrane receptor, "comD", thereby phosphorylating the response regulator, "comE". The phosphorylated comE will activate numerous vital biological mechanisms in Streptococcus mutans such as biofilm formation and the release of mutacin, an antibiotic peptide. Compared with all the other mechanisms involved with CSP, it is found that the promoter of gene "nlmC"( non-lantibiotic mutacin C) in S. mutans has the best performance under the stimulation of CSP.
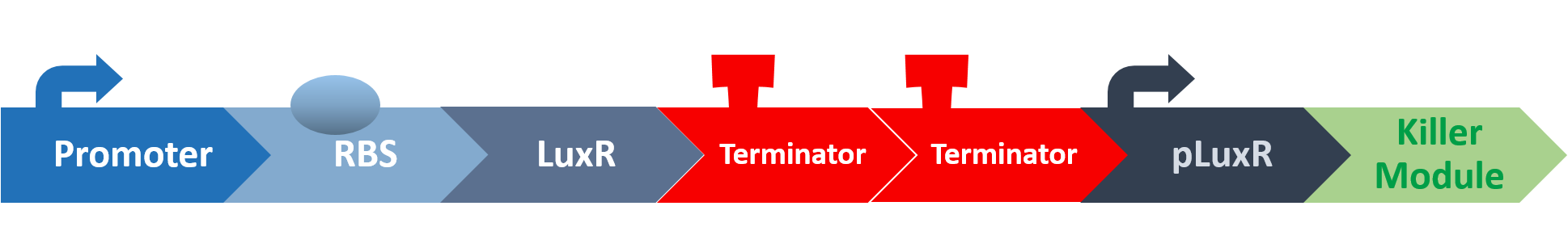
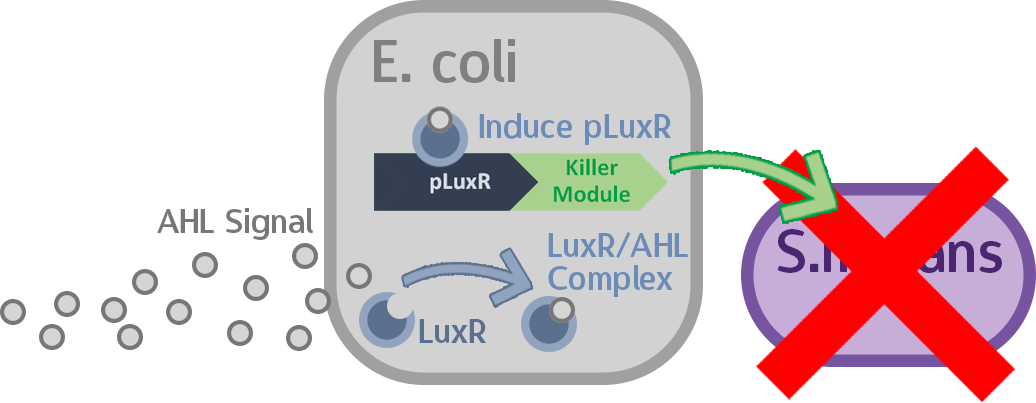
At this point, the two main organisms we adapted for our project are our probiotics and M102 bacteriophage. We used one of the most commonly seen quorum sensing chemicals, AHL, N-Acyl-homoserine-lactone, as our indicator between phage and E.coli. Due to the idea of phage threshold control of the number of S. mutans in prevention of the second-dominant strain in the oral cavity, we envision a possibility to trigger another killing system to reach synergistic effect on diminishing S. mutans. If the threshold terminators were passed, the downstream signaling pathway would be switched on and contact with the engineered probiotics.
When the amount of the S. mutans is so large that the phages couldn't afford the loading, CSP would pass the threshold value and turn on the luxR-luxI system. LuxI acts as a gene which would generate AHL-synthase that yielding the quorum-sensing signal AHL right after the threshold level of S. mutans is exceeded. LuxR is a protein which forms a complex with AHL, and then turns into an activated form which will afterwards bind with the promoter pLuxR and trigger it.
The signal sequence and endolysin downstream would be secreted and eliminate the excess S. mutans.

Figure 1. Gram positive bacteria have only one layer of cell wall outside of cell membrane, composing of peptidoglycan, while the cell wall of gram negative bacteria has additional protection outside peptidoglycan cell wall.[1]
Endolysin is an enzyme expressed by phage-infected bacteria. The C-terminus bind to the host cell wall, while the N-terminus is enzymatic domain. Working with holin, which lyses cell membrane, endolysin could break the cell wall of bacteria. Usually serve as peptidase, endolysin tend to work on gram positive species, which have layers of peptidoglycan cell wall. Though originally intended to be utilized inside the cell wall, endolysin can also be applied outside cell according to previous research. Furthermore, it is even less harmful to gram negative bacteria this way because of the lipid layer of those bacteria composes cell wall.
So how did we do it?

In our circuit, we incorporate nlmC promoter and different combination of terminators to create the threshold.
When the amount of S. mutans is so large that the phages couldn't afford the loading, CSP would pass the threshold value and turn on the luxR-luxI system. LuxI in S. mutans will be triggered by nlmC and activate AHL which will combine with LuxR produced by probiotics continuously. When LuxR combine with AHL to form AHL-LuxR complex, it will induce pLuxR promoter and trigger the Killer Module.
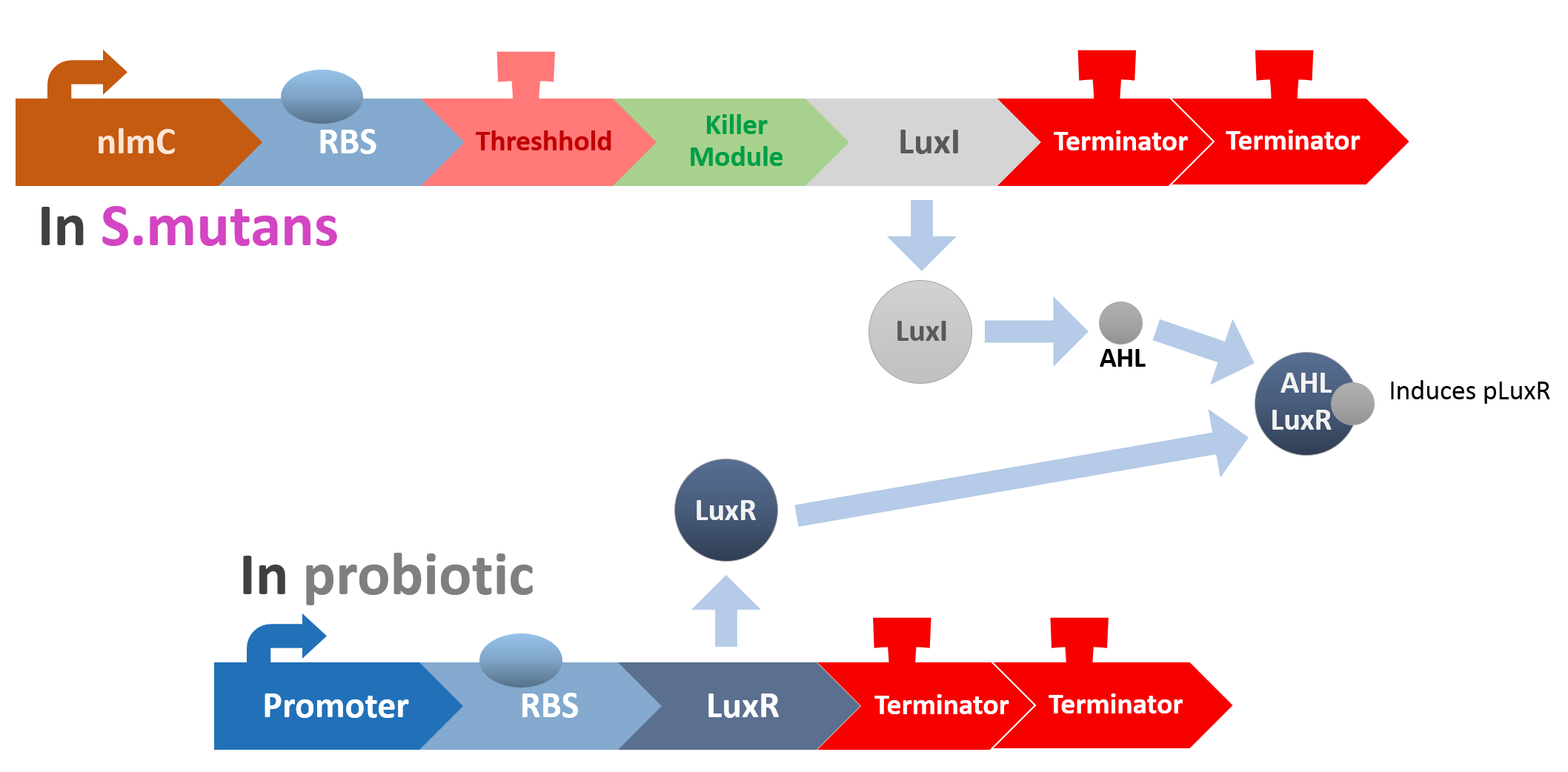
Killer Module
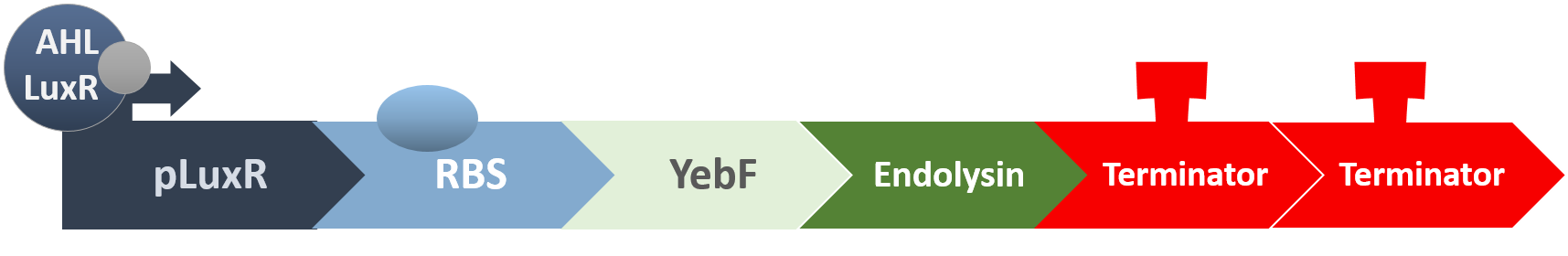
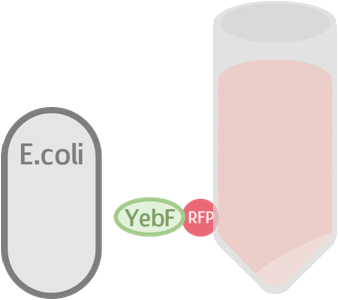
YebF is a protein that is naturally secreted by E.coli. It would transport proteins attached to it to the periplasm (space between cell membrane and cell wall), and later secrete it through porin on the cell wall. We chose YebF for its two advantages: one, it can be secreted in experimental strains which usually do not secrete proteins, along with other diverse passenger characters. Two, YebF can prevent endolysin from toxicating E. coli.
Putting it to the test!
Stage one: How well does nlmC function?
In order to test if nlmC functions as we expected it to, we constructed two circuits. The first is constructed with a constitutive promoter and GFP, and will serve as our control:

The other is constructed instead with nlmC and GFP, and will serve as the testing circuit for comparison between the efficacy of a constitutive promoter and the nlmC promoter:

To compare these circuits, we applied different amounts of synthetic CSP in different time lapses to our experimental group. If everything works as planned, the experimental group should display fluorescence in a wide array of intensity depending on CSP concentration, while the control group displays a uniform bright green.
Stage two: Which threshold combination should we use?
After confirming that the nlmC promoter works and responds to different levels of CSP, we need to design a test to determine which terminator combination we can best use as an effective threshold. We designed two circuits with different terminator thresholds, B0012 and double B0012, respectively, and GFP after them as a reporter.

The testing is similar to Stage 1, and we simply recorded the GFP production after induction with commercial CSP.
In the future, this protocol can be used to test many other combinations to find the "perfect" threshold for HOPE. After obtaining some experimental data, we employed extensive modeling to incorporate these numbers into other combinations of terminators, and we’ll be looking to validate these results after the Jamboree is over to further strengthen HOPE. Please see our [Model: Threshold] section for more information.
Stage three: Mechanisms of the Signal Receptor in our Probiotic
After all the fragments of the circuit is linked, we need a test to tell us that the fragments are well-connected, and that the mechanisms linking them has not lost its function. We created a circuit which is identical to our final circuit, except with RFP in place of the killing module:
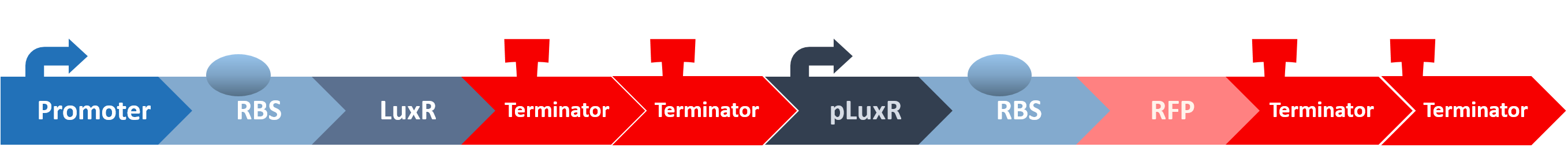
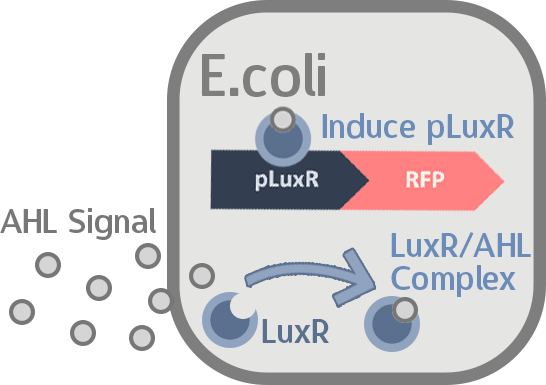
We then transformed our circuit into E. coli, and added in commercial AHL. If the mechanisms remain robust after splicing, we should see a strong display of red from RFP.
Stage four: Mechanisms of the Signal Producer in S. mutans
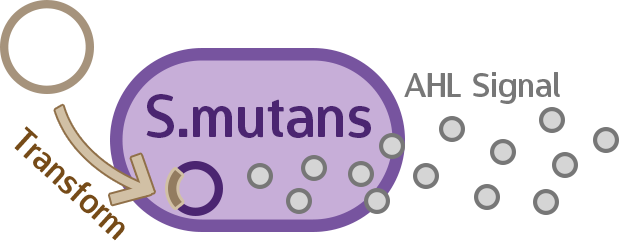
One main concern with our mechanism is that AHL is a Gram-negative quorum sensing-signal… and yet we are implanting it into the Gram-positive S.mutans via M102. We need to test if the AHL production still works in S. mutans.
In order to confirm that whether the engineered circuit could successfully yield AHL, we adopt the purification and structural examination processes referred to related references.The LuxI gene we embedded inside our circuit coded BBa_C0061 in IGEM registry and it synthesizes N-3-(Oxohexanoyl)-L-homoserine lactone. We uses preparative-TLC to purify the chemical and conduct 1HNMR to check the structure.
According to paper research, we prepared our sample through the protocol described below:
- S. mutans liquid cultured till its stationary phase
- Add lysozyme to 5ml cultures
- Sonication
- Centrifugation
- Extract the supernatant with equal volume of ethyl acetate X2
- Dry with anhydrous magnesium sulfate
- Filtering
- Evaporate to dry
- Dissolve with 50-100uL HPLC-grade ethyl acetate
- Run TLC
Stage five: Does YebF work?
Before we check the functionality of the entire construct, we need to test the YebF protein for its ability to transport attached parts out of the membrane. We began by constructing two circuits:
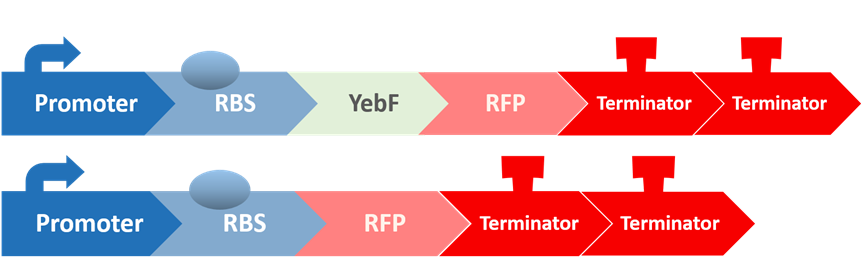
These circuits are then transformed into E. coli, cultivated in liquid culture, and centrifuged. Afterwards, the supernatants are analyzed for flourescence intensity fluorescence intrnsity and their OD value. If YebF works as we expected, we should see the first circuit successfully secrete RFP into the supernatant :
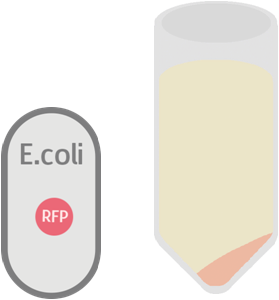
While the control should only contain RFP which is not exported:
Stage six: Does Endolysin work?
To test the antimicrobial activity of endolysin, we also created two circuits:
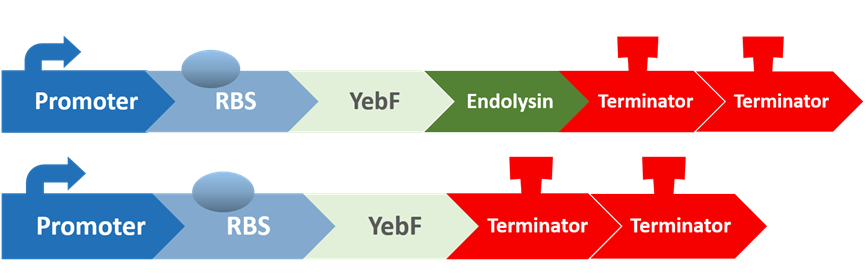
The first circuit contains YebF and endolysin, which serves as the experimental circuit, and the second contains just YebF, and is the control. These are transformed into E. coli, and after culturing them overnight and extracting the supernatant, we spread the supernatant on plates of S. mutans to test for their antimicrobial activity.
Stage seven: Does AHL stimulate the killing program as anticipated?
In this stage, we put the final probiotic circuit into E. coli to see how well it performs.
The two different germs lines are involved in this test, the E.coli constructed circuit (pSB1C3 + luxR + pluxR + YebF + Endolysin) and S.mutans constructed circuit (luxI).
The culturing of constructed E.coli with the supernatant of constructed S.mutans (containing AHL) supposed to be secretion of endolysin. Then we cultured the wild type S.mutans with the supernatant of previous culture.
Stage eight: Finally, let’s check Everything!
In this last stage we put all the constructed circuits intended for our probiotic and M102 into E. coli and S. mutans, respectively. Then we co-cultured these two transformed strains together to test our full circuit.
Our results
nlmC promoter
- Constitutive promoter+ RBS+ GFP
- nlmC promoter+ RBS+ GFP
- nlmC promoter+ RBS+B0012+GFP
- nlmC promoter+ RBS+ B0012+B0012+GFP
Study done by Liu et al. shown that nlmC promoter can be induced by CSP[1]

First of all, “constitutive promoter+ RBS+ GFP” and “nlmC promoter+ RBS+ GFP” will be tested by observing the green fluorescence in the gene-modified Streptococcus mutans. “Constitutive promoter+ RBS+ GFP” is believed to create green fluorescence constitutively without addition stimulation. On the other hand, “nlmC promoter+ RBS +GFP” is believed to create green fluorescence under the stimulation of CSP. In this step of function test, we are hoping to test the function of nlmC promoter.
Secondly, “nlmC promoter+ RBS+ GFP”, “nlmC promoter+ RBS +B0012+ GFP”, and “nlmC promoter +RBS+ B0012+ B0012+GFP” will be tested to see which create the most suitable threshold for preventing cavities. It is believed that the more terminators we added to our circuit, the higher threshold will be made. Under the addition of CSP, the nlmC promoter will be induced, thereby create the green fluorescence which we can observe in the gene-modified Streptococcus mutans.
Testing YebF
From left to right: J23100+yebF+RFP+B0015, J23100, Competent cell

Partially enlargement:

We can see that although there is small difference, there are mostly more fluorescence in our circuit, showing that there might be little RFP secreted. But the effect is not very clear, resulting in small difference in fluorescent intensity.
Also, the comparison between our testing circuit with other control shows that there might exist some technical problems, for sometimes LB or competent cell result in more intensity in RFP wavelength range.
At the same time, measuring OD600 to make comparison :
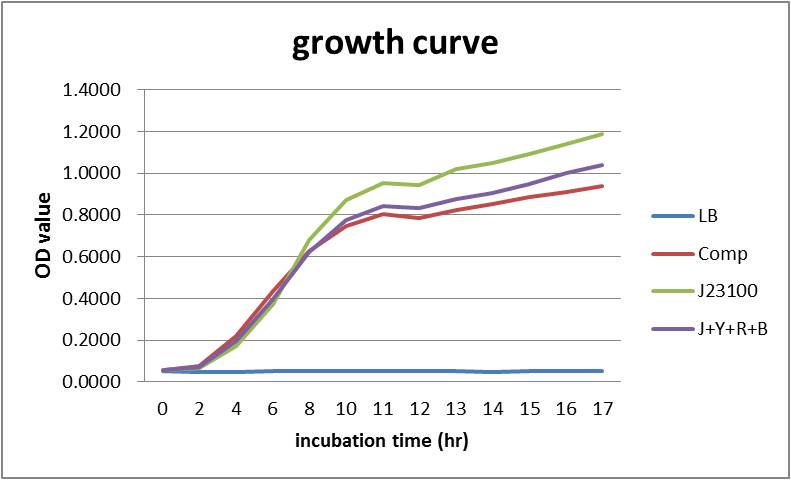
Overall our testing circuit showed the relationship between growth and its secretion as shown below:
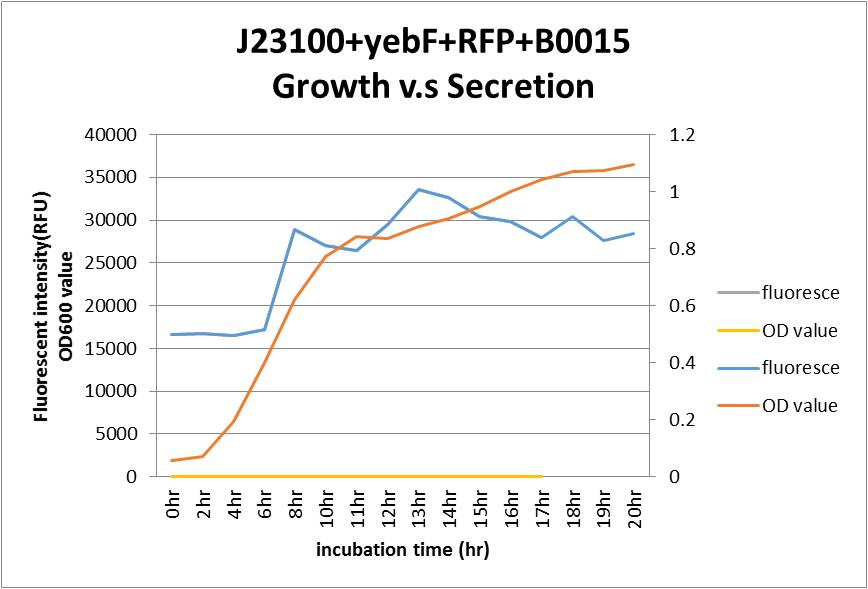
As seen in the graph, the secretion of YebF and RFP basically soared with the growth of the E.coli, and come to a dynamic balance at the as it entered log phase.
Testing Endolysin
By doing disc plate assay of endolysin together with several other controls, we can see that there is a clear space besides the disc of our different concentration of our endolysin supernatant.
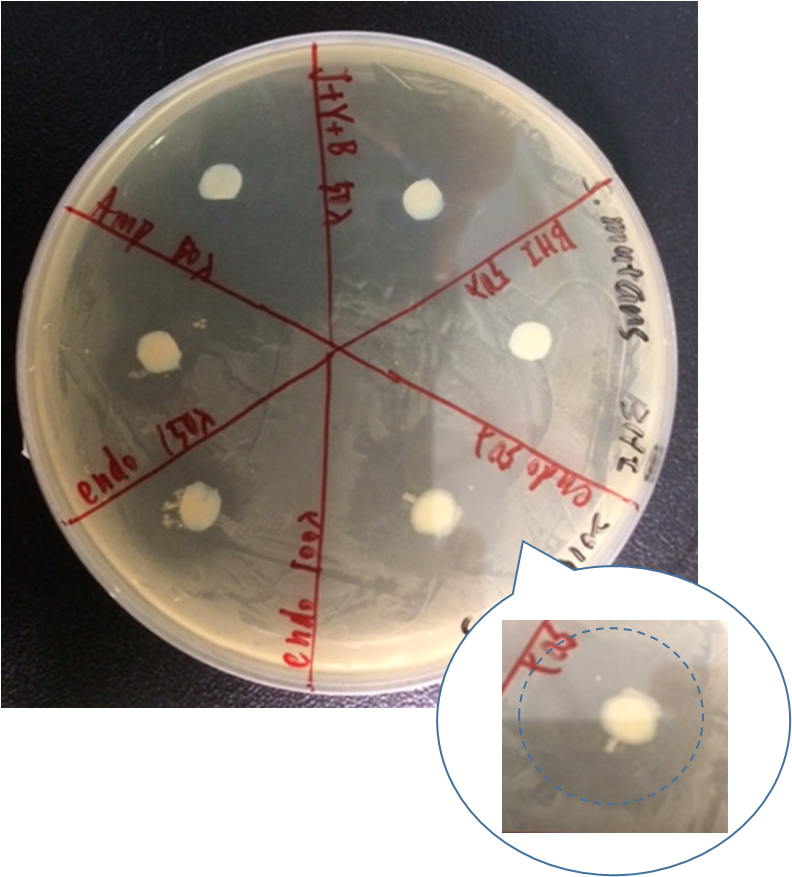
Here are our samples:
- Ampicillin : 100ng/ul
- BHI broth
- J+Y+B: J23100+yebF+B0015 overnight culture supernatant
- (OD600=2.647)(2.0 rcf, 7min)
- J23100+yebF+endolysin+B0015:overnight culture supernatant
- (OD600=2.900)(2.0 rcf, 7min)
Western blot filter paper:
- Positive control: Ampicillin (100ng/ul),50ul
- Negative control: BHI, 50ml /J23100+yebF+B0015, 50ul
- Testing:
- J23100+yebF+endolysin+B0015, 50ul
- J23100+yebF+endolysin+B0015, 100ul
- J23100+yebF+endolysin+B0015, 150ul
As shown in the figure, ampicillin showed a significant inhibition zone as a positive control. Three positive control all show there ability to kill Streptococcus mutans by the clear zone around the disc. The BHI negative control showed no killing ability as expected.
However, there are also little Streptococcus mutans growing near J23100+yebF+B0015 culture supernatant. By observing the pattern on the plate, we speculated that it might be the result of the diffusion of Ampicillin beside it.
AHL induced S. mutans death

The supernatant purified from the S. mutans (with constructed circuit: S-IT/PVA838) contains AHL that could possible trigger the killer circuit in E. coli and show a decreasing growth of wild type S. mutans.
Our constructed circuit (Termed E-C1) including E. coli promoter (Truncated BBa_K523013) region, luxR(BBa_C0062) coding sequence and pLuxR(BBa_R0062) promoter region would be 3117 bp in length. The result of constructed circuit ligation is shown below.
Reference
- Liu, T., et al., ComCED signal loop precisely regulates nlmC expression in Streptococcus mutans. Annals of Microbiology, 2014. 64(1): p. 31-38.
- (2006)"Extracellular accumulation of recombinant proteins fused to the carrier protein YebF in Escherichia coli"
- (2007)"ACES™ Signal Sequence and YebF Expression Systems Technical Brief"
- (2012)"A Protein Export Pathway Involving Escherichia coli Porins"
- (2006)patent" PROTEIN PRODUCTION METHOD UTILIZING YEBF "
- (2007)"Genome sequence of Streptococcus mutans bacteriophage M102"
- (2012)''Biology and Genome Sequence of Streptococcus mutans Phage M102AD''
























 "
"




















