Team:Colombia/Stochastic
From 2014.igem.org
| Line 17: | Line 17: | ||
[[File:Colombia_PetriNet.png|center|600px]] | [[File:Colombia_PetriNet.png|center|600px]] | ||
| + | |||
| + | <i> | ||
| + | “…For “ordinary” chemical systems in which fluctuations and correlations play no significant role, the method stands as an alternative to the traditional procedure of numerically solving the deterministic reaction rate equations. For nonlinear systems near chemical instabilities, where fluctuations and correlations may invalidate the deterministic equations, the method constitutes an efficient way of numerically examining the predictions of the stochastic master equation. Although fully equivalent to the spatially homogeneous master equation, the numerical…” (Gillespie 1976) | ||
| + | |||
| + | </i> | ||
Revision as of 21:19, 17 October 2014
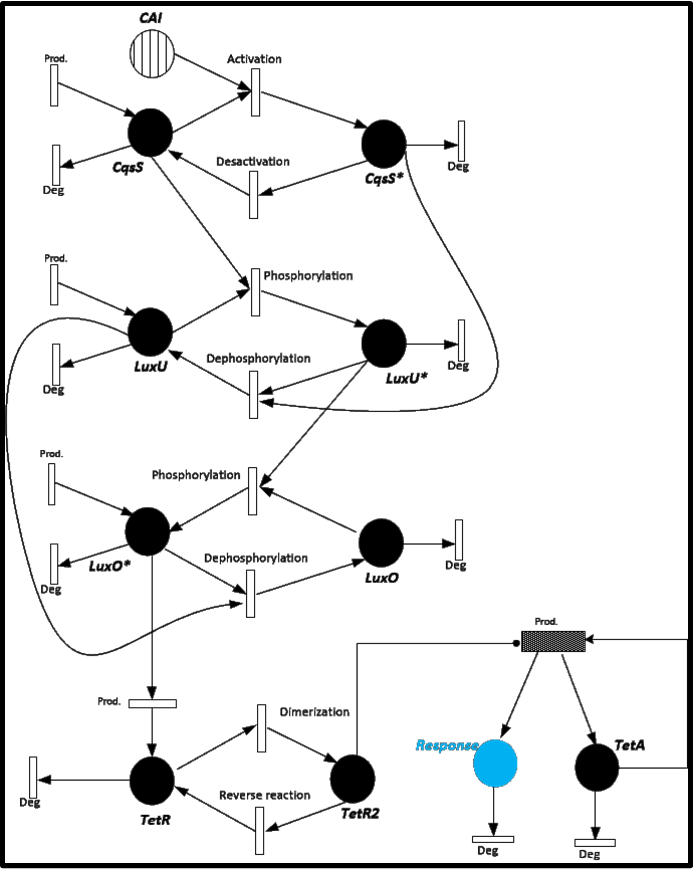
Stochastic Model.
Until now, all we have is a deterministic model of our system of detection of Cholerae. This model takes into account exact concentrations of molecules as variables in the differential equations. This means that we make calculations with the mean values of the amount of molecules.
It’s well known that we can simulate this kind of molecular dynamics as stochastic processes. In particular, such an events will we simulated as Markovian random walk in N dimension, where N represents the number of substances involved.
“…For “ordinary” chemical systems in which fluctuations and correlations play no significant role, the method stands as an alternative to the traditional procedure of numerically solving the deterministic reaction rate equations. For nonlinear systems near chemical instabilities, where fluctuations and correlations may invalidate the deterministic equations, the method constitutes an efficient way of numerically examining the predictions of the stochastic master equation. Although fully equivalent to the spatially homogeneous master equation, the numerical…” (Gillespie 1976)
 "
"