Team:Paris Saclay/Notebook/August/27
From 2014.igem.org
(→Wednesday 27th August) |
m (→Photo of the Day) |
||
| (26 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
=Wednesday 27th August= | =Wednesday 27th August= | ||
==Lab Work== | ==Lab Work== | ||
| - | === | + | ===Construction of the fusion protein (color)=== |
| + | |||
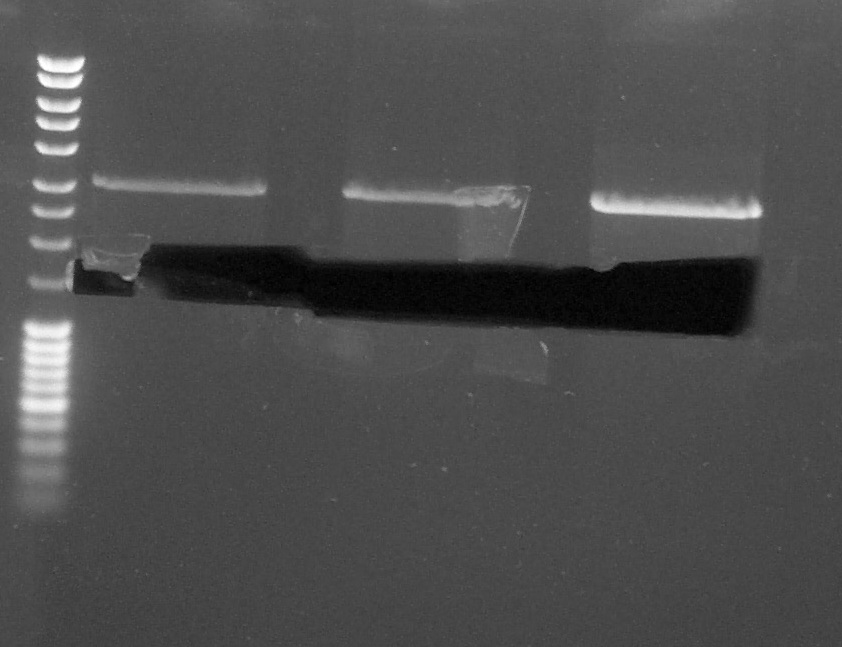
| + | ==== Electrophoresis of pGEMTeasy+chromoprotein 3,4 and 6 by EcoRI and PstI ==== | ||
| + | [[File:270814 Laetitia gel decoup chromo.jpg|300px|left]] | ||
| + | ''by Laetitia'' | ||
| + | |||
| + | Digestion made the 26th by Laetitia. After the migration on agarose gel (100V), we cut the band corresponding to the chromoprotein gene on UV table. | ||
| + | |||
| + | |||
| + | |||
| + | ====Purification of the chromoprotein gene from the agarose gel ==== | ||
| + | ''by Sean'' | ||
| + | |||
| + | We used the kit PCR-Clean-UP to extract the DNA from the agarose gel. Elution volume : 15µl | ||
| + | |||
| + | ====Gel electrophoresis of the purification product ==== | ||
| + | ''by Sean'' | ||
| + | |||
| + | The goal is to check if the purification of the chromoprotein gene has worked. | ||
| + | |||
| + | PCR clean-up was performed ealier [https://2014.igem.org/Team:Paris_Saclay/Notebook/August/27#PCR_clean-up_of_colonies_containing_pGEMTeasy.2Bchromoprotein today]. 1µl of each result was used for the electrophoresis. Nothing appeared under UV. | ||
| + | |||
| + | ====Ligation of chromoprotein==== | ||
| + | ''by Sean'' | ||
| + | |||
| + | {| class="wikitable centre" width="20%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | components | ||
| + | ! scope=col | volumes | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |6μl | ||
| + | |- | ||
| + | |T4 DNA ligase buffer | ||
| + | |2µl | ||
| + | |- | ||
| + | |purified chromoprotein | ||
| + | |10µl | ||
| + | |- | ||
| + | |ligase | ||
| + | |1µl | ||
| + | |- | ||
| + | |pSB1C3 | ||
| + | |1µl | ||
| + | |} | ||
| + | |||
| + | Leave overnight at 16°C. | ||
| + | |||
| + | ====Collection of pellets containing pGEMTeasy+chromoprotein==== | ||
| + | ''by Sean'' | ||
| + | |||
| + | Three liquid cultures (pGEM+chromoprotein) were prepared. 2ml from each were collected in 2ml microcentrifuge tubes, then centrifuged. The pellets were put in the freezer. | ||
| + | |||
| + | ==== Transformation of odor free E. coli with plasmids coding Fluo Protein ==== | ||
| + | ''by Terry'' | ||
| + | |||
| + | Our fusion protein was not expressed in pGEMTeasy (no color in the culture), so, if our system do not work at all, I'm preparing FP (Fluorescent Protein) for our lemon. | ||
| + | |||
| + | 6 plamids coding FP have been transformed in Odor Free : | ||
| + | *pCFP+ | ||
| + | *pYFP+ | ||
| + | *pRFP+ | ||
| + | *pEGFP | ||
| + | *pGFP | ||
| + | *pEYFP | ||
| + | |||
| + | ===lemon scent === | ||
====Plasmide extraction==== | ====Plasmide extraction==== | ||
'' By Mélanie'' | '' By Mélanie'' | ||
| Line 8: | Line 75: | ||
Extraction of pPS5 with the phenol protocol (as previously described) | Extraction of pPS5 with the phenol protocol (as previously described) | ||
| + | ====PCR of Limonene synthase (BBa762100)==== | ||
| + | ''by Mélanie'' | ||
| + | |||
| + | 5 tubes to do a gradient | ||
| + | |||
| + | {| class="wikitable centre" width="50%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | components | ||
| + | ! scope=col | volumes | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |29.75μl | ||
| + | |- | ||
| + | |Gotaq buffer | ||
| + | |10µl | ||
| + | |- | ||
| + | |dNTP 10mM | ||
| + | |1µl | ||
| + | |- | ||
| + | |iPS67 | ||
| + | |1µM | ||
| + | |- | ||
| + | |iPS66 | ||
| + | |1µM | ||
| + | |- | ||
| + | |DNA | ||
| + | |1µl | ||
| + | |- | ||
| + | |MgCl2 | ||
| + | |4µl | ||
| + | |- | ||
| + | |Gotaq | ||
| + | |1.25µl | ||
| + | |} | ||
| + | |||
| + | {| class="wikitable centre" width="50%" | ||
| + | |+ PCR cycle | ||
| + | |- | ||
| + | ! scope=col | step | ||
| + | ! scope=col | temperature (°C) | ||
| + | ! scope=col | time | ||
| + | |- | ||
| + | |1 | ||
| + | |95 | ||
| + | |2min | ||
| + | |- | ||
| + | |2 | ||
| + | |95 | ||
| + | |30 sec | ||
| + | |- | ||
| + | |3 | ||
| + | |49-55 (gradient) | ||
| + | |30sec | ||
| + | |- | ||
| + | |4 | ||
| + | |72 | ||
| + | |2min | ||
| + | |- | ||
| + | |5 | ||
| + | |72 | ||
| + | |5min | ||
| + | |} | ||
| + | |||
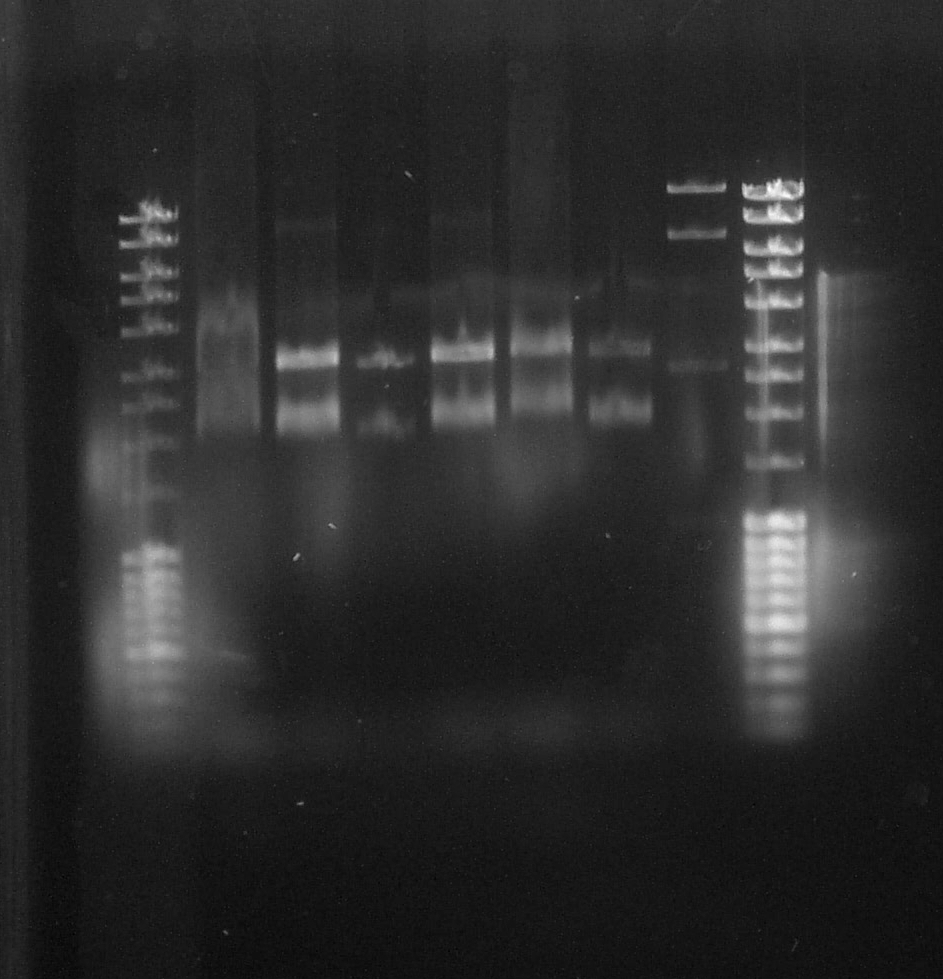
| + | ==== Electrophoresis of pPS3 and pPS4==== | ||
| + | |||
| + | [[File:Paris_Saclay_electrophoresis_pPS3_pPS4.jpeg|400px|right]] | ||
| + | |||
| + | ''by Terry'' | ||
| + | |||
| + | Result from [https://2014.igem.org/Team:Paris_Saclay/Notebook/August/26 yesterday] 's extraction. | ||
| + | |||
| + | Gel shaft : | ||
| + | |||
| + | *0 Ladder | ||
| + | *1 pPS4 (geraniol synthase) | ||
| + | *2 pPS3 (pinene synthase) | ||
| + | *3 pPS3 | ||
| + | *4 pPS4 | ||
| + | *5 pPS4 | ||
| + | *6 pPS4 | ||
| + | *7 pPS4 | ||
| + | *8 Ladder | ||
| + | |||
| + | |||
| + | The pPS4 (shaft 4) seems to correspond to our reference but the gel is not top quality. More analysis need to be made. | ||
| + | |||
| + | |||
| + | ==Photo of the Day== | ||
| + | [[File:Paris Saclay 27_august.jpg|600px|center]] | ||
| + | |||
| + | '''Members present:''' | ||
| + | * Instructors and advisors: Alice, Solenne and Sylvie. | ||
| + | * Students: Eugène, Hoang Vu, Laëtitia, Mélanie, Romain, Sean and Terry. | ||
{{Team:Paris_Saclay/notebook_footer}} | {{Team:Paris_Saclay/notebook_footer}} | ||
Latest revision as of 15:20, 14 October 2014
Wednesday 27th August
Lab Work
Construction of the fusion protein (color)
Electrophoresis of pGEMTeasy+chromoprotein 3,4 and 6 by EcoRI and PstI
by Laetitia
Digestion made the 26th by Laetitia. After the migration on agarose gel (100V), we cut the band corresponding to the chromoprotein gene on UV table.
Purification of the chromoprotein gene from the agarose gel
by Sean
We used the kit PCR-Clean-UP to extract the DNA from the agarose gel. Elution volume : 15µl
Gel electrophoresis of the purification product
by Sean
The goal is to check if the purification of the chromoprotein gene has worked.
PCR clean-up was performed ealier today. 1µl of each result was used for the electrophoresis. Nothing appeared under UV.
Ligation of chromoprotein
by Sean
| components | volumes |
|---|---|
| H2O | 6μl |
| T4 DNA ligase buffer | 2µl |
| purified chromoprotein | 10µl |
| ligase | 1µl |
| pSB1C3 | 1µl |
Leave overnight at 16°C.
Collection of pellets containing pGEMTeasy+chromoprotein
by Sean
Three liquid cultures (pGEM+chromoprotein) were prepared. 2ml from each were collected in 2ml microcentrifuge tubes, then centrifuged. The pellets were put in the freezer.
Transformation of odor free E. coli with plasmids coding Fluo Protein
by Terry
Our fusion protein was not expressed in pGEMTeasy (no color in the culture), so, if our system do not work at all, I'm preparing FP (Fluorescent Protein) for our lemon.
6 plamids coding FP have been transformed in Odor Free :
- pCFP+
- pYFP+
- pRFP+
- pEGFP
- pGFP
- pEYFP
lemon scent
Plasmide extraction
By Mélanie
Extraction of pPS5 with the phenol protocol (as previously described)
PCR of Limonene synthase (BBa762100)
by Mélanie
5 tubes to do a gradient
| components | volumes |
|---|---|
| H2O | 29.75μl |
| Gotaq buffer | 10µl |
| dNTP 10mM | 1µl |
| iPS67 | 1µM |
| iPS66 | 1µM |
| DNA | 1µl |
| MgCl2 | 4µl |
| Gotaq | 1.25µl |
| step | temperature (°C) | time |
|---|---|---|
| 1 | 95 | 2min |
| 2 | 95 | 30 sec |
| 3 | 49-55 (gradient) | 30sec |
| 4 | 72 | 2min |
| 5 | 72 | 5min |
Electrophoresis of pPS3 and pPS4
by Terry
Result from yesterday 's extraction.
Gel shaft :
- 0 Ladder
- 1 pPS4 (geraniol synthase)
- 2 pPS3 (pinene synthase)
- 3 pPS3
- 4 pPS4
- 5 pPS4
- 6 pPS4
- 7 pPS4
- 8 Ladder
The pPS4 (shaft 4) seems to correspond to our reference but the gel is not top quality. More analysis need to be made.
Photo of the Day
Members present:
- Instructors and advisors: Alice, Solenne and Sylvie.
- Students: Eugène, Hoang Vu, Laëtitia, Mélanie, Romain, Sean and Terry.
 "
"