Team:Paris Saclay/Notebook/August/25
From 2014.igem.org
(→Electrophoresis of the digestion product of pGEMTeasy+chromoprotein) |
m (→Monday 25th August) |
||
| (19 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
==Lab work== | ==Lab work== | ||
| - | === | + | ===Construction of the fusion protein=== |
====plasmid extraction : pGEMTeasy + chromoprotein ==== | ====plasmid extraction : pGEMTeasy + chromoprotein ==== | ||
| Line 48: | Line 48: | ||
T : control negatif without enzyme | T : control negatif without enzyme | ||
| - | + | {| class="wikitable centre" width="50%" | |
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | component | ||
| + | ! scope=col | volume | ||
| + | |- | ||
| + | |blue | ||
| + | |2 μl | ||
| + | |- | ||
| + | |Plasmid | ||
| + | |1 μl | ||
| + | |- | ||
| + | |H20 | ||
| + | |9μl | ||
| + | |} | ||
| - | |||
| - | [[File:250814 laetitia digestion verif chromoP pGEMTeasy.jpg| | + | [[File:250814 laetitia digestion verif chromoP pGEMTeasy.jpg|400px|right]] |
| - | Results : the samples 3, 4 and 6 seems to contain the chromoprotein gene (two bands) | + | D : sample digested by XbaI and PstI |
| + | |||
| + | T1 - D1 - T2 - D2 - T3 - D3 - E - T4 - D4 - T5 - o - D6 | ||
| + | |||
| + | |||
| + | Results : lost of the sample D5 in a hole in the agarose gel | ||
| + | |||
| + | The samples 3, 4 and 6 seems to contain the chromoprotein gene (two bands) | ||
| + | |||
| + | --> We will try another plasmid extraction because the electrophoresis profil doesn't show bands for every sample. | ||
==== Bacterial culture of DH5a containing pGEMTeasy + chromprotein ==== | ==== Bacterial culture of DH5a containing pGEMTeasy + chromprotein ==== | ||
| Line 60: | Line 82: | ||
We launched 6 cultures in 5mL LB + Ampi (1/1000) - 37°C at night . The bacteria come from the stock made before the plasmid extraction | We launched 6 cultures in 5mL LB + Ampi (1/1000) - 37°C at night . The bacteria come from the stock made before the plasmid extraction | ||
| - | |||
===D - Lemon Scent=== | ===D - Lemon Scent=== | ||
====Gel electrophoresis of CAD==== | ====Gel electrophoresis of CAD==== | ||
| + | |||
| + | [[File:Paris_Saclay_Sean_140825.jpg|left]] | ||
| + | |||
''by Sean'' | ''by Sean'' | ||
| + | |||
| + | Veirfication of the purification from [https://2014.igem.org/Team:Paris_Saclay/Notebook/August/22#PCR_of_CAD Friday] | ||
''Legend'' | ''Legend'' | ||
| Line 70: | Line 96: | ||
# PCR result of CAD | # PCR result of CAD | ||
| - | [[File: | + | |
| + | |||
| + | ==== CAD ligation in pPS1 ==== | ||
| + | ''by Mélanie'' | ||
| + | pPSI was previously digest by PacI and dephosphorylated | ||
| + | We want to insert the CAD PCR fragment in pPSI to make pPS5. | ||
| + | |||
| + | {| class="wikitable centre" width="50%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | component | ||
| + | ! scope=col | volume | ||
| + | |- | ||
| + | |PCR CAD | ||
| + | |10 μl | ||
| + | |- | ||
| + | |pPS1 | ||
| + | |3μl | ||
| + | |- | ||
| + | |10x buffer ligase | ||
| + | |2μl | ||
| + | |- | ||
| + | |ligase | ||
| + | |1μl | ||
| + | |- | ||
| + | |H20 | ||
| + | |4μl | ||
| + | |} | ||
| + | |||
| + | 4H at room temperature | ||
| + | |||
| + | ==== Transformation of DH5a with the ligation ==== | ||
| + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/Transformation_of_supercompetent_Ecoli_cells_with_CaCl2 protocol] | ||
| + | |||
| + | |||
| + | ====Culture of bacteria with pPS2 - pPS3 - pPS4==== | ||
| + | I take some colonies made [https://2014.igem.org/Team:Paris_Saclay/Notebook/August/22#Transformation_in_competent_E.coli_DH5.CE.B1 Friday 22th August] | ||
| + | |||
| + | === Limonene synthase digestion=== | ||
| + | |||
| + | Because we don't success to have pPS2 - we try to digest again Limonene synthase in pGMET esay | ||
| + | |||
| + | {| class="wikitable centre" width="25%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | components | ||
| + | ! scope=col | volumes | ||
| + | |- | ||
| + | |LS pGMET esay | ||
| + | |50μl | ||
| + | |- | ||
| + | |FastDigest green buffer 10X | ||
| + | |5μl | ||
| + | |- | ||
| + | |PacI | ||
| + | |0.5µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |4.5µl | ||
| + | |} | ||
| + | |||
| + | when I done the electrophoresis to check the digestion, I have a surprise : no digestion. | ||
| + | |||
| + | So we check the pGMETesay plasmid with normaly the inserts (for LS GS and PS)and we saw that there is no insert in the plasmid. | ||
| + | |||
| + | |||
| + | ==Photo of the Day== | ||
| + | [[File:Paris Saclay 25_august.jpg|600px|center]] | ||
| + | |||
| + | '''Members present:''' | ||
| + | * Instructors and advisors: Alice, Solenne and Sylvie. | ||
| + | * Students: Eugène, Hoang Vu, Laëtitia, Mélanie, Romain, Sean and Terry. | ||
{{Team:Paris_Saclay/notebook_footer}} | {{Team:Paris_Saclay/notebook_footer}} | ||
Latest revision as of 15:14, 14 October 2014
Contents |
Monday 25th August
Lab work
Construction of the fusion protein
plasmid extraction : pGEMTeasy + chromoprotein
by Laetitia
We used the bacteria containing pGEMTeasy + chromoprotein launched by Melanie sunday the 24th for the plasmid extraction, 6 independent cultures which come from 6 independent colonies.
We made a stock of bacteria for each sample (x6).
To extract the plasmid, we used the plasmid DNA purification kit (Macherey-Nagel).
Digestion of pGEMTeasy+chromoproteinby XbaI and PstI
by Laetitia
The goal is to check the presence of the insert inside the plasmid. We digest the 6 samples of purified plasmid.
| component | volume |
|---|---|
| Plasmid | 5 μl |
| Fast Digest buffer 10X | 1μl |
| XbaI | 0.5μl |
| PstI | 0.5μl |
| H20 | 3μl |
1h at 37°C
Electrophoresis of the digestion product of pGEMTeasy+chromoprotein
by Laetitia
T : control negatif without enzyme
| component | volume |
|---|---|
| blue | 2 μl |
| Plasmid | 1 μl |
| H20 | 9μl |
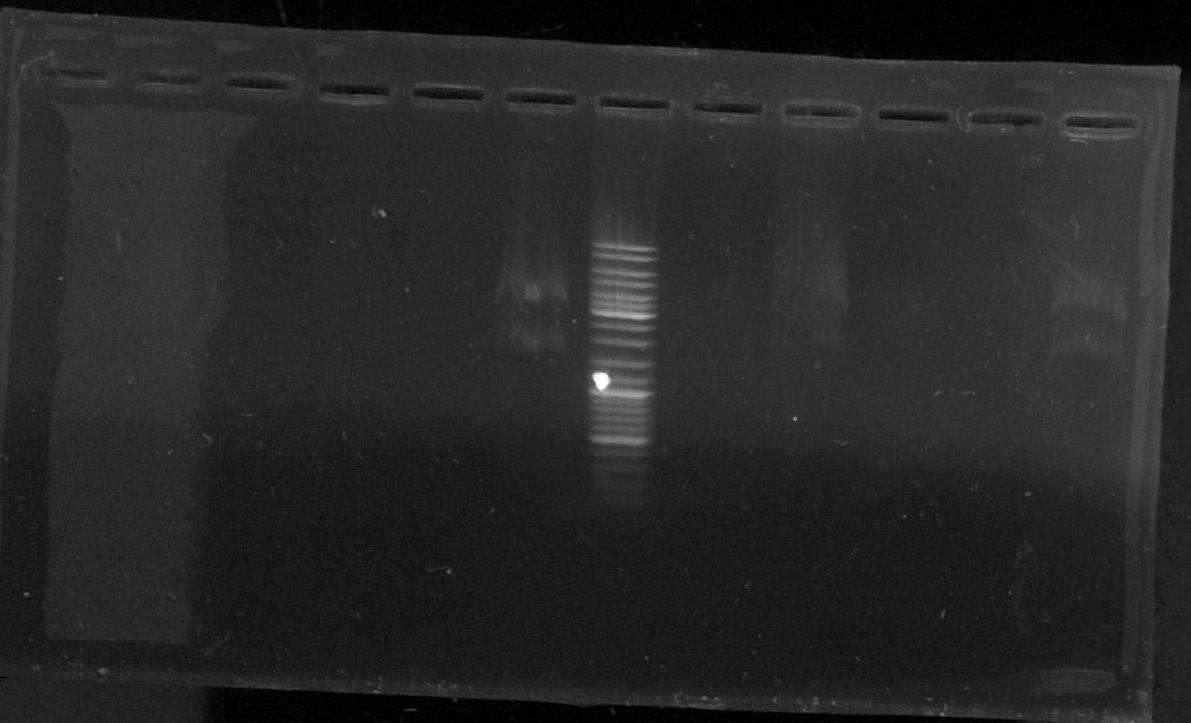
D : sample digested by XbaI and PstI
T1 - D1 - T2 - D2 - T3 - D3 - E - T4 - D4 - T5 - o - D6
Results : lost of the sample D5 in a hole in the agarose gel
The samples 3, 4 and 6 seems to contain the chromoprotein gene (two bands)
--> We will try another plasmid extraction because the electrophoresis profil doesn't show bands for every sample.
Bacterial culture of DH5a containing pGEMTeasy + chromprotein
by Laetitia
We launched 6 cultures in 5mL LB + Ampi (1/1000) - 37°C at night . The bacteria come from the stock made before the plasmid extraction
D - Lemon Scent
Gel electrophoresis of CAD
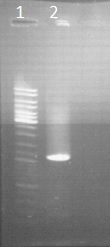
by Sean
Veirfication of the purification from Friday
Legend
- ladder 10µl
- PCR result of CAD
CAD ligation in pPS1
by Mélanie pPSI was previously digest by PacI and dephosphorylated We want to insert the CAD PCR fragment in pPSI to make pPS5.
| component | volume |
|---|---|
| PCR CAD | 10 μl |
| pPS1 | 3μl |
| 10x buffer ligase | 2μl |
| ligase | 1μl |
| H20 | 4μl |
4H at room temperature
Transformation of DH5a with the ligation
Culture of bacteria with pPS2 - pPS3 - pPS4
I take some colonies made Friday 22th August
Limonene synthase digestion
Because we don't success to have pPS2 - we try to digest again Limonene synthase in pGMET esay
| components | volumes |
|---|---|
| LS pGMET esay | 50μl |
| FastDigest green buffer 10X | 5μl |
| PacI | 0.5µl |
| H2O | 4.5µl |
when I done the electrophoresis to check the digestion, I have a surprise : no digestion.
So we check the pGMETesay plasmid with normaly the inserts (for LS GS and PS)and we saw that there is no insert in the plasmid.
Photo of the Day
Members present:
- Instructors and advisors: Alice, Solenne and Sylvie.
- Students: Eugène, Hoang Vu, Laëtitia, Mélanie, Romain, Sean and Terry.
 "
"