Team:Paris Saclay/Notebook/August/11
From 2014.igem.org
Terry.Mara (Talk | contribs) (→PCR Clean-up of BBa_K762100) |
m (→Photo of the Day) |
||
| (36 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{Team:Paris_Saclay/notebook_header}} | ||
=Monday 11th August= | =Monday 11th August= | ||
==Lab work== | ==Lab work== | ||
| - | === | + | |
| + | ===The chassis coli Odor free=== | ||
| + | |||
| + | ''by Melanie'' | ||
| + | |||
| + | Preculture odor free | ||
| + | |||
| + | ===Salicylate Inducible Suppressing System=== | ||
| + | ====Liquid Culture==== | ||
| + | ''by Fabio'' | ||
| + | |||
| + | 4 liquid cultures of '''BBa_K1372001''' with 20ml of LB and 20µl of antibiotic Cm (at 5pm - 150 rpm - 37 °C), from the stock made the [https://2014.igem.org/Team:Paris_Saclay/Notebook/August/8#Transformation_of_competent_E.coli_cells 8th August]. | ||
| + | |||
| + | ===Lemon scent=== | ||
====PCR Clean-up of BBa_K762100==== | ====PCR Clean-up of BBa_K762100==== | ||
''by Sean'' | ''by Sean'' | ||
| - | PCR | + | PCR prepared on the [https://2014.igem.org/Team:Paris_Saclay/Notebook/August/7#Polymerase_chain_reaction 7th August]. |
| + | |||
| + | Clean-up performed with the following protocol. | ||
| + | |||
| + | <span style="color:red">NB: in the present case we have 90 µl of PCR result and we use 20 µl of elution buffer.</span> | ||
| + | |||
| + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/PCR_clean-up Protocol] | ||
| + | |||
| + | ====Digestion==== | ||
| + | |||
| + | ''by Laetitia and Hoang Vu'' | ||
| + | |||
| + | ====Check the replacement of Limonen synthase gene by Apramycin resistant gene in the pJBEI plasmid.==== | ||
| + | |||
| + | 1)3 µl plasmid pJBEI (or plasmid pJBEI+ApraR 2) | ||
| + | 0.5 µl BglII | ||
| + | 1 µl fast digest buffer | ||
| + | 10 µl H<sub>2</sub>O QSP | ||
| + | |||
| + | 37°C - 1 hour | ||
| + | Migration on agarose gel | ||
| + | |||
| + | ====Check if the restriction enzymes PacI from Sylvie's and from Alice's stock work.==== | ||
| + | |||
| + | 1) 3 µl plasmid pJBEI (or plasmid pJBEI+ApraR 2) | ||
| + | 0.5 µl PacI(Alice) | ||
| + | 1 µl PacI buffer | ||
| + | 10 µl H<sub>2</sub>O QSP | ||
| + | |||
| + | 2)3 µl plasmid pJBEI (or plasmid pJBEI+ApreR 2) | ||
| + | 0.5 µl PacI(Sylvie) | ||
| + | 1 µl PacI buffer | ||
| + | 10 µl H<sub>2</sub>O QSP | ||
| + | |||
| + | 37°C - 1 hour | ||
| + | Migration on agarose gel | ||
| + | |||
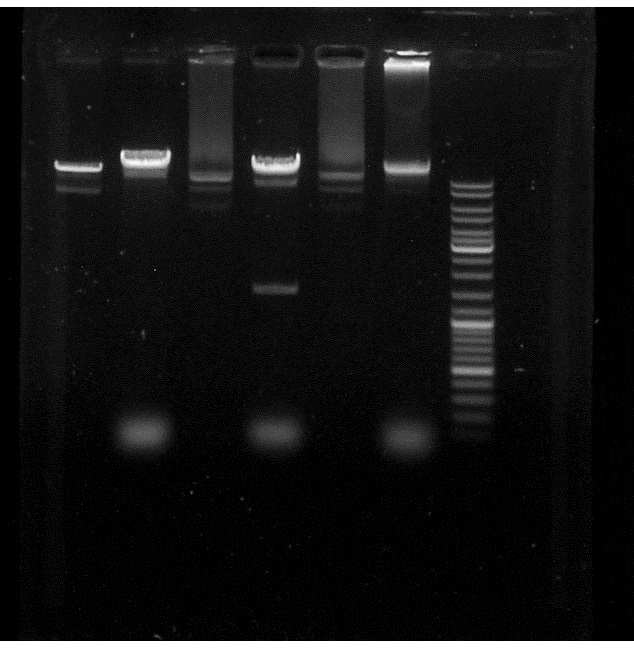
| + | '''Conclusion:''' | ||
| + | Alice's PacI works because we can see a supplementary band in the pJBEI+ApraR 2 compared to the control but not the Sylvie's. | ||
| + | So, we do have the ApraR gene in our plasmid. | ||
| + | |||
| + | |||
| + | [[File:PacI_et_BglII_restriction_Apra_in_pJBEI.jpg]] | ||
| + | |||
| + | ====Transformation of E.coli DH5 α by pJBEI+ApraR 2==== | ||
| + | |||
| + | 100µl competent bacteria | ||
| + | 2 µl pJBEI+Apra 2 | ||
| + | |||
| + | 20'' at 4°C | ||
| + | 2''30' at 42°C | ||
| + | 2'' at 4°C | ||
| + | |||
| + | Then, we add 900µL of lysogeny broth and put it | ||
| + | |||
| + | 1 hour at 37°C | ||
| + | |||
| + | In parallel, we did a control. (Without adding plasmid) | ||
| + | |||
| + | Then, we spread our ''E.coli'' on the petri dish containing Solid LB+Apra (1/2000) | ||
| + | 100 µl control | ||
| + | 100 µl transformed ''E.coli'' | ||
| + | 200 µl transformed ''E.coli'' | ||
| + | 100 µl concentrated transformed ''E.coli'' | ||
| + | |||
| + | '''Results:''' | ||
| + | Nothing grew. | ||
| + | We think that the problem comes from the competent DH5 α that were not really competent... | ||
| + | Edit : bacteria grew but 48h after the culture | ||
| + | |||
| + | ==Human Practices== | ||
| + | ===Art & Design=== | ||
| + | ''by Terry'' | ||
| + | |||
| + | Incubation of a preculture of ''E. Coli'' with plasmid FNR RBS AmylCP (BB used in iGEM 2013 Paris-Saclay) coding for a blue | ||
| + | chromoprotein. Will be used tomorrow for a test of colored bacteria density in agar. | ||
| + | |||
| + | ==Photo of the Day== | ||
| + | [[File:Paris Saclay 11_august.jpg|600px|center]] | ||
| + | |||
| + | '''Members present:''' | ||
| + | * Instructors and advisors: Alice, Solenne and Sylvie. | ||
| + | * Students: Eugène, Fabio, Hoang Vu, Mélanie, Romain Sean and Terry. | ||
| + | |||
| + | {{Team:Paris_Saclay/notebook_footer}} | ||
Latest revision as of 14:51, 14 October 2014
Monday 11th August
Lab work
The chassis coli Odor free
by Melanie
Preculture odor free
Salicylate Inducible Suppressing System
Liquid Culture
by Fabio
4 liquid cultures of BBa_K1372001 with 20ml of LB and 20µl of antibiotic Cm (at 5pm - 150 rpm - 37 °C), from the stock made the 8th August.
Lemon scent
PCR Clean-up of BBa_K762100
by Sean
PCR prepared on the 7th August.
Clean-up performed with the following protocol.
NB: in the present case we have 90 µl of PCR result and we use 20 µl of elution buffer.
Digestion
by Laetitia and Hoang Vu
Check the replacement of Limonen synthase gene by Apramycin resistant gene in the pJBEI plasmid.
1)3 µl plasmid pJBEI (or plasmid pJBEI+ApraR 2) 0.5 µl BglII 1 µl fast digest buffer 10 µl H2O QSP
37°C - 1 hour Migration on agarose gel
Check if the restriction enzymes PacI from Sylvie's and from Alice's stock work.
1) 3 µl plasmid pJBEI (or plasmid pJBEI+ApraR 2) 0.5 µl PacI(Alice) 1 µl PacI buffer 10 µl H2O QSP
2)3 µl plasmid pJBEI (or plasmid pJBEI+ApreR 2) 0.5 µl PacI(Sylvie) 1 µl PacI buffer 10 µl H2O QSP
37°C - 1 hour Migration on agarose gel
Conclusion: Alice's PacI works because we can see a supplementary band in the pJBEI+ApraR 2 compared to the control but not the Sylvie's. So, we do have the ApraR gene in our plasmid.
Transformation of E.coli DH5 α by pJBEI+ApraR 2
100µl competent bacteria 2 µl pJBEI+Apra 2
20 at 4°C 230' at 42°C 2 at 4°C
Then, we add 900µL of lysogeny broth and put it
1 hour at 37°C
In parallel, we did a control. (Without adding plasmid)
Then, we spread our E.coli on the petri dish containing Solid LB+Apra (1/2000) 100 µl control 100 µl transformed E.coli 200 µl transformed E.coli 100 µl concentrated transformed E.coli
Results: Nothing grew. We think that the problem comes from the competent DH5 α that were not really competent... Edit : bacteria grew but 48h after the culture
Human Practices
Art & Design
by Terry
Incubation of a preculture of E. Coli with plasmid FNR RBS AmylCP (BB used in iGEM 2013 Paris-Saclay) coding for a blue chromoprotein. Will be used tomorrow for a test of colored bacteria density in agar.
Photo of the Day
Members present:
- Instructors and advisors: Alice, Solenne and Sylvie.
- Students: Eugène, Fabio, Hoang Vu, Mélanie, Romain Sean and Terry.
 "
"