Team:Paris Saclay/Notebook/August/5
From 2014.igem.org
(→Miscellaneous) |
m (→Tuesday 5th August) |
||
| (26 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
=Tuesday 5th August= | =Tuesday 5th August= | ||
==Lab work== | ==Lab work== | ||
| - | === | + | ===The chassis coli Odor free=== |
| + | ====PCR verification==== | ||
| + | ''by Romain'' | ||
| + | |||
| + | Results of the striates on dishes from [https://2014.igem.org/Team:Paris_Saclay/Notebook/August/4 yesterday]: | ||
| + | |||
| + | For the strain MG1655Z1: | ||
| + | *Nothing has grown on the kan dish, but the colony grew on LB dish. That was what we expected, the kan cassette was correctly delete. | ||
| + | |||
| + | For the strain MG1655: | ||
| + | *Few colonies have grown on the kan dish, and all the colonies have grown on LB dish. It's a little failure. | ||
| + | |||
| + | So now, we proceed to the PCR to verify the length of the sequences. | ||
| + | |||
| + | Strains used: E. coli MG1655Z1 and E. coli MG1655, and oligonucleotides used: iPS75 bis and iPS76 bis. | ||
| + | |||
| + | '''Protocol''' | ||
| + | |||
| + | Add into a PCR tube the following: | ||
| + | |||
| + | {| class="wikitable centre" width="50%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | Component | ||
| + | ! scope=col | For a total volume of 50μl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |28.25μl | ||
| + | |- | ||
| + | | Green GoTaq buffer 5X | ||
| + | | 10μl | ||
| + | |- | ||
| + | | dNTPs 10mM | ||
| + | | 1μl | ||
| + | |- | ||
| + | | iPS75 bis 10µM | ||
| + | | 2μl | ||
| + | |- | ||
| + | | iPS76 bis 10µM | ||
| + | | 2μl | ||
| + | |- | ||
| + | | DMSO | ||
| + | | 1.5μl | ||
| + | |- | ||
| + | | MgCl<sub>2</sub> 25mM | ||
| + | | 4μl | ||
| + | |- | ||
| + | | Bacterial culture | ||
| + | | 2μl | ||
| + | |- | ||
| + | | Green GoTaq enzyme | ||
| + | | 0.25μl | ||
| + | |} | ||
| + | |||
| + | We follow these quantities for 11 tubes: 5 tubes for the strain MG1655, 5 tubes for the strain MG1655Z1 and 1 tube with bacteria which had grown on the kan dish. | ||
| + | Tube was placed in PCR machine with the following parameters. | ||
| + | |||
| + | {| class="wikitable centre" width="80%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | Cycle step | ||
| + | ! scope=col | Temperature | ||
| + | ! scope=col | Time | ||
| + | ! scope=col | Cycle | ||
| + | |- | ||
| + | | width="25%" | | ||
| + | Initial denaturation | ||
| + | | width="25%" | | ||
| + | 95°C | ||
| + | | width="25%" | | ||
| + | 10 min | ||
| + | | width="25%" | | ||
| + | | | ||
| + | |- | ||
| + | | Denaturation | ||
| + | | 95°C | ||
| + | | 30 s | ||
| + | | 30 | ||
| + | |- | ||
| + | | Annealing | ||
| + | | 53°C | ||
| + | | 30 s | ||
| + | | 30 | ||
| + | |- | ||
| + | | Extension | ||
| + | | 72°C | ||
| + | | 2 min | ||
| + | | 30 | ||
| + | |- | ||
| + | | Final extension | ||
| + | | 72°C | ||
| + | | 5 min | ||
| + | | 1 | ||
| + | |- | ||
| + | | Final extension | ||
| + | | 12°C | ||
| + | | hold | ||
| + | | 1 | ||
| + | |} | ||
| + | |||
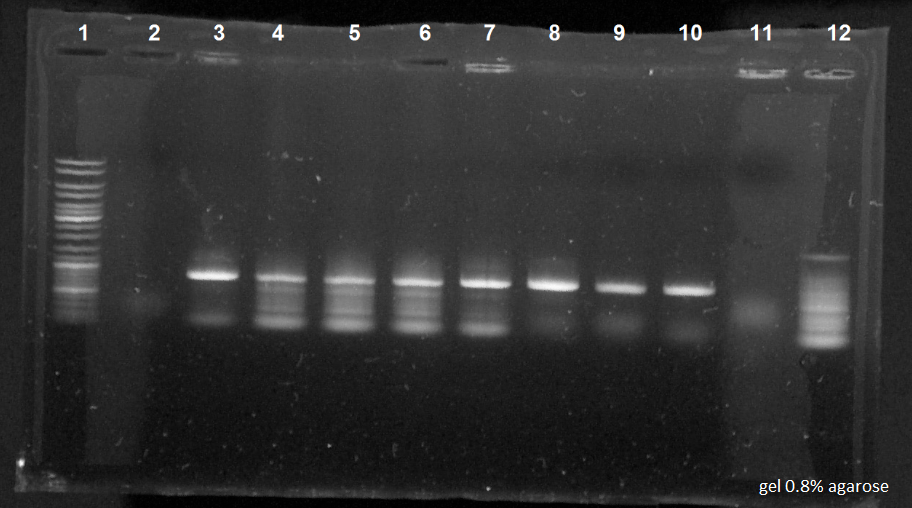
| + | The different bacterial cultures in each tubes: | ||
| + | [[File:050814-Romain-PCRa.jpg|400px|right]] | ||
| + | *Tube 1 to 5: MG1655 | ||
| + | *Tube 6 to 10: MG1655Z1 | ||
| + | *Tube 11: MG1655 which had grown on the kan dish. | ||
| + | |||
| + | The electrophoresis results: | ||
| + | *1. 10 µl of ladder | ||
| + | *2 to 6. 10 µl in each tube of PCR for the strain MG1655 | ||
| + | *7 to 11. 10 µl in each tube of PCR for the strain MG1655Z1 | ||
| + | *12. 10µl of PCR for the MG1655 which had grown on the kan dish | ||
| + | |||
| + | Results of the PCR: The electrophoresis revealed correctly the expected size of the sequence, except for the holes 2 and 11. | ||
| + | The kan resistance sequence had been successfully removed. | ||
| + | |||
| + | Liquid cultures overnight made: 3ml LB medium + 500µl transformed strain MG1655 + Cm | ||
| + | |||
| + | ===Salicylate Inducible Suppressing System=== | ||
| + | ====Liquid Culture==== | ||
| + | ''by Fabio'' | ||
| + | |||
| + | 2 liquid cultures of '''BBa_K1372000''' with 20ml of LB and 20µl of amp (at 3pm - 150 rpm - 37 °C), from the stock made the [https://2014.igem.org/Team:Paris_Saclay/Notebook/August/4#Final_Stock 4th August]. | ||
| + | |||
| + | ===Lemon scent=== | ||
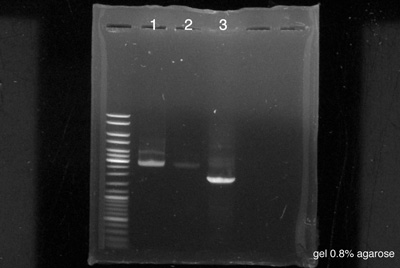
====Electrophoresis==== | ====Electrophoresis==== | ||
[[File:050814-Sean-Pierre-PCRa.jpg|400px|right]] | [[File:050814-Sean-Pierre-PCRa.jpg|400px|right]] | ||
| Line 13: | Line 136: | ||
[https://2014.igem.org/Team:Paris_Saclay/Protocols/Gel_Electrophoresis Protocol] | [https://2014.igem.org/Team:Paris_Saclay/Protocols/Gel_Electrophoresis Protocol] | ||
| - | + | ==Human Practices== | |
| - | + | ||
| - | ==Human | + | |
''by Pierre'' | ''by Pierre'' | ||
| Line 30: | Line 151: | ||
*Bichat, Physiological researches on life and death | *Bichat, Physiological researches on life and death | ||
| + | ''by Hoang Vu'' | ||
| + | |||
| + | Continue my bibliographical research on Citral A and B and Beta-Pinene. | ||
| + | I have now a little more than twenty reviews about various uses and properties of citral | ||
| + | |||
| + | ===Art & Design=== | ||
| + | ''by Leila & Juliette'' | ||
| + | |||
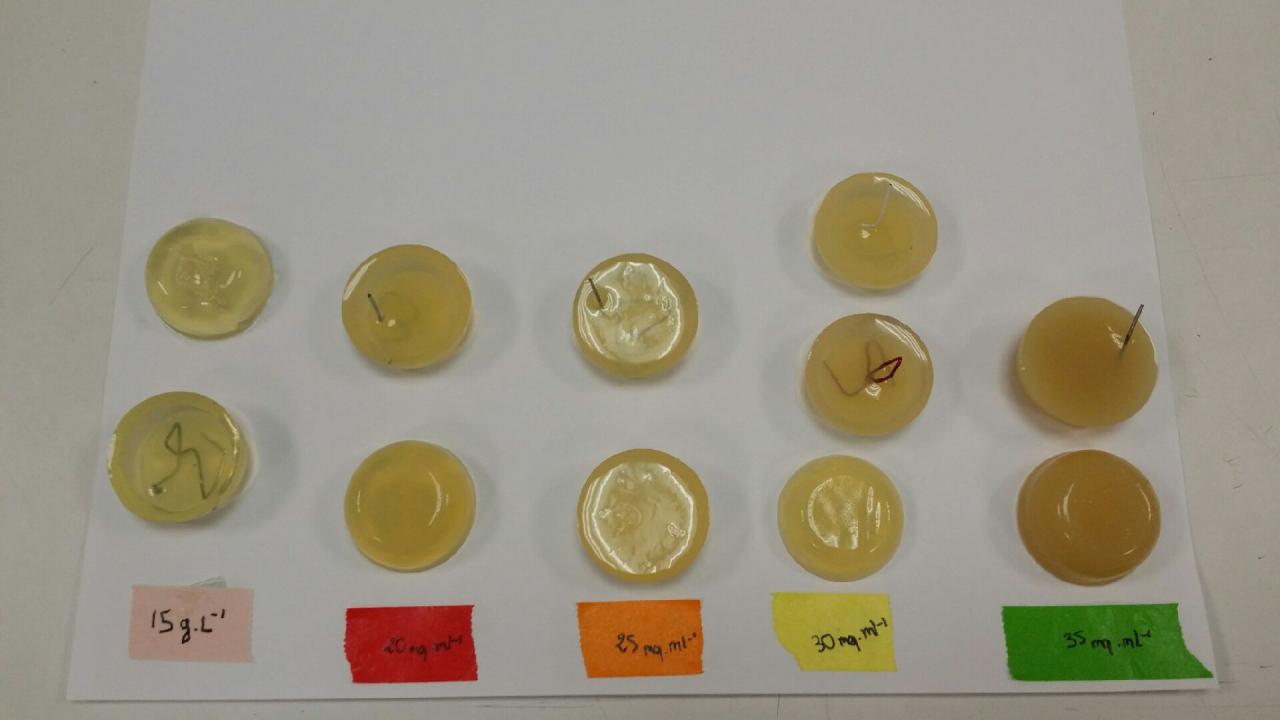
| + | [[File:Test1.jpg|400px|left]] | ||
| + | |||
| + | We tested some different concentration of agar in LAG, how we can hang it, how it looks and how easy it is to turn it out : 15 g/l, 20 g/l, 25 g/l, 30 g/l, 35 g/l. | ||
| + | |||
| + | {| class="wikitable centre" width="50%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | Concentration | ||
| + | ! scope=col | Strength | ||
| + | ! scope=col | Color | ||
| + | |- | ||
| + | |15 g/l | ||
| + | |to much liquid : we can't hang it | ||
| + | |really translucent | ||
| + | |- | ||
| + | | 20 g/l | ||
| + | | good holding and resistance | ||
| + | | quite translucent | ||
| + | |- | ||
| + | | 25 g/l | ||
| + | | good holding and resistance | ||
| + | | quite translucent | ||
| + | |- | ||
| + | | 30 g/l | ||
| + | | good holding and resistance | ||
| + | | not so much translucent | ||
| + | |- | ||
| + | | 35 g/l | ||
| + | | really resistant. Maybe a little bit too heavy. | ||
| + | | opaque | ||
| + | |} | ||
| + | |||
| + | |||
| + | Finally, we though our test were effective and we decided to select only 3 concentration for the future : 20 g/l, 25 g/l and 30 g/l. | ||
| + | |||
| + | [[File:Test2.jpg|500px|center]] | ||
| + | |||
| + | ==Photo of the Day== | ||
| + | [[File:Paris Saclay 5_august.jpg|600px|center]] | ||
'''Members there''': | '''Members there''': | ||
| - | * Instructors and advisors: Alice | + | * Instructors and advisors: Alice and Sylvie. |
* Students: Eugene, Fabio, Hoang Vu, Leila, Juliette, Pierre, Romain, Sean and Terry. | * Students: Eugene, Fabio, Hoang Vu, Leila, Juliette, Pierre, Romain, Sean and Terry. | ||
| - | + | {{Team:Paris_Saclay/notebook_footer}} | |
| - | + | ||
Latest revision as of 14:22, 14 October 2014
Contents |
Tuesday 5th August
Lab work
The chassis coli Odor free
PCR verification
by Romain
Results of the striates on dishes from yesterday:
For the strain MG1655Z1:
- Nothing has grown on the kan dish, but the colony grew on LB dish. That was what we expected, the kan cassette was correctly delete.
For the strain MG1655:
- Few colonies have grown on the kan dish, and all the colonies have grown on LB dish. It's a little failure.
So now, we proceed to the PCR to verify the length of the sequences.
Strains used: E. coli MG1655Z1 and E. coli MG1655, and oligonucleotides used: iPS75 bis and iPS76 bis.
Protocol
Add into a PCR tube the following:
| Component | For a total volume of 50μl |
|---|---|
| H2O | 28.25μl |
| Green GoTaq buffer 5X | 10μl |
| dNTPs 10mM | 1μl |
| iPS75 bis 10µM | 2μl |
| iPS76 bis 10µM | 2μl |
| DMSO | 1.5μl |
| MgCl2 25mM | 4μl |
| Bacterial culture | 2μl |
| Green GoTaq enzyme | 0.25μl |
We follow these quantities for 11 tubes: 5 tubes for the strain MG1655, 5 tubes for the strain MG1655Z1 and 1 tube with bacteria which had grown on the kan dish. Tube was placed in PCR machine with the following parameters.
| Cycle step | Temperature | Time | Cycle | |
|---|---|---|---|---|
|
Initial denaturation |
95°C |
10 min | ||
| Denaturation | 95°C | 30 s | 30 | |
| Annealing | 53°C | 30 s | 30 | |
| Extension | 72°C | 2 min | 30 | |
| Final extension | 72°C | 5 min | 1 | |
| Final extension | 12°C | hold | 1 |
The different bacterial cultures in each tubes:
- Tube 1 to 5: MG1655
- Tube 6 to 10: MG1655Z1
- Tube 11: MG1655 which had grown on the kan dish.
The electrophoresis results:
- 1. 10 µl of ladder
- 2 to 6. 10 µl in each tube of PCR for the strain MG1655
- 7 to 11. 10 µl in each tube of PCR for the strain MG1655Z1
- 12. 10µl of PCR for the MG1655 which had grown on the kan dish
Results of the PCR: The electrophoresis revealed correctly the expected size of the sequence, except for the holes 2 and 11. The kan resistance sequence had been successfully removed.
Liquid cultures overnight made: 3ml LB medium + 500µl transformed strain MG1655 + Cm
Salicylate Inducible Suppressing System
Liquid Culture
by Fabio
2 liquid cultures of BBa_K1372000 with 20ml of LB and 20µl of amp (at 3pm - 150 rpm - 37 °C), from the stock made the 4th August.
Lemon scent
Electrophoresis
by Sean & Pierre The strains used were made the 4th August
- 5 µl of BBa_K762100 non diluted
- 5 µl of BBa_K762100 diluted 10-1
- 5 µl of BBa_K517003
Human Practices
by Pierre
Ethics
I wrote the begining of a global view of the evolution of the definitions of life from Aristotle to modern times.
Bibliography
- Aristotle, On the Soul
- Epicurus, Letter to Herodotus
- Descartes, Discourse on the method
- Darwin, On the origin of species
- Bichat, Physiological researches on life and death
by Hoang Vu
Continue my bibliographical research on Citral A and B and Beta-Pinene. I have now a little more than twenty reviews about various uses and properties of citral
Art & Design
by Leila & Juliette
We tested some different concentration of agar in LAG, how we can hang it, how it looks and how easy it is to turn it out : 15 g/l, 20 g/l, 25 g/l, 30 g/l, 35 g/l.
| Concentration | Strength | Color |
|---|---|---|
| 15 g/l | to much liquid : we can't hang it | really translucent |
| 20 g/l | good holding and resistance | quite translucent |
| 25 g/l | good holding and resistance | quite translucent |
| 30 g/l | good holding and resistance | not so much translucent |
| 35 g/l | really resistant. Maybe a little bit too heavy. | opaque |
Finally, we though our test were effective and we decided to select only 3 concentration for the future : 20 g/l, 25 g/l and 30 g/l.
Photo of the Day
Members there:
- Instructors and advisors: Alice and Sylvie.
- Students: Eugene, Fabio, Hoang Vu, Leila, Juliette, Pierre, Romain, Sean and Terry.
 "
"