Team:Paris Saclay/Notebook/August/4
From 2014.igem.org
(→Monday 4th August) |
m (→Monday 4th August) |
||
| (28 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{Team:Paris_Saclay/notebook_header}} | ||
=Monday 4th August= | =Monday 4th August= | ||
| - | |||
==Lab Work== | ==Lab Work== | ||
| - | === | + | ===The chassis coli Odor free=== |
| - | + | ||
====Striate on Dishes==== | ====Striate on Dishes==== | ||
''by Juliette & Romain'' | ''by Juliette & Romain'' | ||
| - | We | + | We screened MG1655 and MG1655Z1 colonies on LB and LB + Kanamycin dishes for the loss of kanamycin resistance cassette, using the colonies obtained from the streak plates prepared on LB dishes and incubated at 42°C. [https://2014.igem.org/Team:Paris_Saclay/Notebook/August/1 1st August] |
| - | === | + | ===Salicylate Inducible Suppressing System=== |
| + | ====Plasmid DNA Purification==== | ||
| + | ''by Eugene and Fabio'' | ||
| + | * Ligation of '''BBa_J61051''' and '''BBa_K228001''' made the [https://2014.igem.org/Team:Paris_Saclay/Notebook/July/30#Ligation 30th July] | ||
| + | From Liquid Culture made the [https://2014.igem.org/Team:Paris_Saclay/Notebook/August/1#Liquid_Culture 1st August] | ||
| + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/Extraction_of_the_Genomic_DNA_from_Bacteria_by_using_NucleoSpin®_Tissue Protocol] | ||
| + | |||
| + | ====Digestion to check==== | ||
| + | ''by Eugene and Fabio'' | ||
| + | |||
| + | In order to validate the hole Assembly Process, we did a final digestion. The samples were digested with the enzymes '''EcoRI''' and '''PstI''', that separates the BioBrick Assembly core from theirs vectors. The graphic result of this process is shown in the next step. | ||
| + | |||
| + | Protocol: | ||
| + | # Add 5 μl of the plasmid to digest. | ||
| + | # Add 2 μl of buffer (Fast Digest Buffer 10x). | ||
| + | # Add 0,2 μl of enzyme EcoRI. | ||
| + | # Add 0,2 μl of enzyme PstI. | ||
| + | # Complete with 12,6μl of H<sub>2</sub>O. | ||
| + | # Mix gently. | ||
| + | # Incubate at 37°C for one hour. | ||
| + | |||
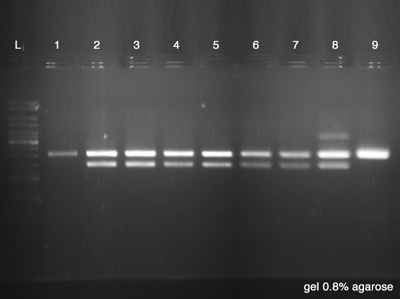
| + | ====Electrophoresis==== | ||
| + | [[File:LU000114a.jpg|400px|right]] | ||
| + | ''by Fabio'' | ||
| + | |||
| + | The Electrophoresis was used to verify the success of the Digestion and the success of the plasmid DNA purification at the same time. Strain number 9 represents the digestion's control as it is the '''BBa_K228001''', the BioBrick used as vector. If the clones have the same size of the control, it indicates that we had no ligation between both BioBricks ('''BBa_J61051''' and '''BBa_K228001'''). ''Each sample has 20 μl and the Ladder has 5 μl.'' | ||
| + | |||
| + | # Plasmid Cl.1 | ||
| + | # Plasmid Cl.2 | ||
| + | # Plasmid Cl.3 | ||
| + | # Plasmid Cl.4 | ||
| + | # Plasmid Cl.5 | ||
| + | # Plasmid Cl.6 | ||
| + | # Plasmid Cl.7 | ||
| + | # Plasmid Cl.8 | ||
| + | # BBa_K228001 - Control | ||
| + | '''Results:''' | ||
| + | # Failed, the plasmid had no digestion. | ||
| + | # Success, the plasmid had the good size of digestion. | ||
| + | # Success, the plasmid had the good size of digestion. | ||
| + | # Success, the plasmid had the good size of digestion. | ||
| + | # Success, the plasmid had the good size of digestion. | ||
| + | # Success, the plasmid had the good size of digestion. | ||
| + | # Success, the plasmid had the good size of digestion. | ||
| + | # Failed, the plasmid had others digestions points. | ||
| + | # Good, we have a good control. | ||
| + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/BioBrick_Assembly#Segregate_process BioBrick Assembly - Segregate Process Protocol] | ||
| + | |||
| + | ====Final Stock==== | ||
| + | ''by Fabio'' | ||
| + | |||
| + | As the BioBrick Assembly was a success, proved by the last Electrophoresis, we have registered a new BioBrick named '''BBa_K1372000''' together with its final stock. We used the Clones 2, 3 and 5 from Liquid Colonies made the [https://2014.igem.org/Team:Paris_Saclay/Notebook/August/1#Liquid_Culture 1st August] as the Electrophoresis shows a good concentration of them. | ||
| + | * BBa_K1372000 Cl. 1 | ||
| + | * BBa_K1372000 Cl. 2 | ||
| + | * BBa_K1372000 Cl. 3 | ||
| + | ''1 ml of colonies plus 0.25 ml of glycerol 87%'' | ||
| + | |||
| + | ===D - Lemon scent=== | ||
====Gel electrophoresis of BBa_K762100==== | ====Gel electrophoresis of BBa_K762100==== | ||
''by Sean'' | ''by Sean'' | ||
| Line 21: | Line 77: | ||
From left to right: 5 µl of BBa_K762100 + "old" Phusion enzyme, 5 µl of BBa_K762100 + "new" Phusion enzyme | From left to right: 5 µl of BBa_K762100 + "old" Phusion enzyme, 5 µl of BBa_K762100 + "new" Phusion enzyme | ||
| - | [https://2014.igem.org/Team:Paris_Saclay/Notebook | + | ====Electroporation==== |
| + | ''by Terry'' | ||
| + | |||
| + | Strain used: DY330 containing plasmid pJBEI6409. | ||
| + | We had to switch the Limonen Synthase cassette with the Apramycin resistance. | ||
| + | |||
| + | 15 ml of culture has been placed at 42°C for 17 min then cooled 10 min before centrifugation. | ||
| + | |||
| + | Make 2 électroporations in cold electroporation cuvettes: | ||
| + | |||
| + | *A control cuvette: 50µl of DY330. | ||
| + | *A second cuvette: 50µl of DY330 culture + 8µl of PCR purified product identified the [https://2014.igem.org/Team:Paris_Saclay/Notebook/July/21 21st July] | ||
| + | |||
| + | Electroporation : 2500V, 132W, 40µF. | ||
| + | |||
| + | After that, add 1ml of cold LB in each cuvette and transfer in tubes to incubate during 1 to 2h at 30°C. | ||
| + | |||
| + | Spread on 4 dishes LB + Apramicine : | ||
| + | |||
| + | *100µl of control | ||
| + | *50µl of transformed DY330 with 8µl of PCR purified product | ||
| + | *100µl of transformed DY330 with 8µl of PCR purified product | ||
| + | *The rest of transformed DY330 with 8µl of PCR purified product | ||
| + | |||
| + | Incubate for 2 days at 30°C. | ||
| + | |||
| + | ==== Polymerase chain reaction ==== | ||
| + | ''by Sean & Pierre'' | ||
| + | |||
| + | '''BBa_K517003''' | ||
| + | |||
| + | {| class="wikitable centre" width="50%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | component | ||
| + | ! scope=col | volume | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |35.5μl | ||
| + | |- | ||
| + | | Phusion buffer 5X | ||
| + | | 10μl | ||
| + | |- | ||
| + | | dNTPs | ||
| + | | 2μl | ||
| + | |- | ||
| + | | iPS68bis | ||
| + | | 1μl | ||
| + | |- | ||
| + | | iPS69 | ||
| + | | 1μl | ||
| + | |- | ||
| + | | DNA | ||
| + | | 1μl | ||
| + | |- | ||
| + | | Phusion enzyme | ||
| + | | 0.5μl | ||
| + | |} | ||
| + | |||
| + | |||
| + | '''BBa_K762100''' | ||
| + | |||
| + | For this BioBrick two tubes were prepared: one with undiluted DNA and one with DNA diluted 10<sup>-1</sup>. | ||
| + | |||
| + | {| class="wikitable centre" width="50%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | component | ||
| + | ! scope=col | volume | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |35.5μl | ||
| + | |- | ||
| + | |Phusion buffer 5X | ||
| + | |10μl | ||
| + | |- | ||
| + | |dNTPs | ||
| + | |2μl | ||
| + | |- | ||
| + | |iPS66 | ||
| + | |1μl | ||
| + | |- | ||
| + | |iPS67 | ||
| + | |1μl | ||
| + | |- | ||
| + | |DNA | ||
| + | |1μl | ||
| + | |- | ||
| + | |Phusion enzyme | ||
| + | |0.5μl | ||
| + | |} | ||
| + | |||
| + | Tubes were placed in PCR machine with the following parameters. | ||
| + | |||
| + | {| class="wikitable centre" width="80%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | Cycle step | ||
| + | ! scope=col | Temperature | ||
| + | ! scope=col | Time | ||
| + | ! scope=col | Cycle | ||
| + | |- | ||
| + | | width="25%" | | ||
| + | Initial denaturation | ||
| + | | width="25%" | | ||
| + | 98°C | ||
| + | | width="25%" | | ||
| + | 1 min | ||
| + | | width="25%" | | ||
| + | 1 | ||
| + | |- | ||
| + | | Denaturation | ||
| + | | 98°C | ||
| + | | 15 s | ||
| + | | 25 - 30 | ||
| + | |- | ||
| + | | Annealing | ||
| + | | 52°C | ||
| + | | 25 s | ||
| + | | 25 - 30 | ||
| + | |- | ||
| + | | Extension | ||
| + | | 72°C | ||
| + | | 45 s | ||
| + | | 25-30 | ||
| + | |- | ||
| + | | Final extension | ||
| + | | 72°C | ||
| + | | 10 min | ||
| + | | 1 | ||
| + | |- | ||
| + | | Final extension | ||
| + | | 8°C | ||
| + | | hold | ||
| + | | 1 | ||
| + | |} | ||
| + | |||
| + | ===Gel electrophoresis of BBa_K517003 and K762100 previously prepared=== | ||
| + | |||
| + | by Sean&Pierre | ||
| + | |||
| + | |||
| + | Results | ||
| + | |||
| + | failure. | ||
| + | |||
| + | From left to right: 10 µl ladder, 5 µl of BBa_K762100 non diluted, 5 µl of BBa_K762100 diluted 10<sup>-1</sup>, 5 µl of BBa_K517003. | ||
| + | |||
| + | ==Miscellaneous== | ||
| + | |||
| + | By Hoang Vu | ||
| + | |||
| + | ===Ethics=== | ||
| + | Well, I just got back from one month of vacation and when I came, everybody was already on lab works. So, I decided to work on ethics first. | ||
| + | Synthetic Biology and BioArt are controversial because one is about engineering living organisms, and therefore raises economic, religious, political, moral problems; and the other is about engineering the living organisms too but it's mostly USING the living organisms to do art, to carry a message. This is how I see the essential points of our project ethical problems. | ||
| + | But my point of view is not fixed. Maybe I'm wrong, maybe there is something that I have missed. That's why I started with a research on GMOs in general to better understand those issues. | ||
| + | * The first trangenic pet were GloFish, "TK-1" 2003 in Taiwan | ||
| + | * California is completely against such animals but the funny thing is that California is one of the best supporters of agricultural GMOs. So, there is the question: Why the plants and not the fish? Probably economic benefits and ethical positions are the reasons. Then, is there a hierachy of living organisms? Why? | ||
| + | * GMOs under commercial cover raise new problems on environment and commercial rights. | ||
| + | * Take a look at the arguments between Anti-GMO and Pro-GMO movements. The 90's context for example in Europe after food crises like the mad cow disease, contribute to the incomplete acceptance by the Europeans. | ||
| + | |||
| + | ===Bibliography=== | ||
| + | I worked on ethics till one of our advisors ask me: "Can you do a research on the chemical components of the lemon odor please? I need more publications on chemical characterization of limonene, citral A and B, and beta-Pinene" | ||
| + | |||
| + | |||
| + | ==Photo of the Day== | ||
| + | [[File:Paris Saclay 4_august.jpg|600px|center]] | ||
| + | |||
| + | '''Members there''': | ||
| + | * Instructors and advisors: Alice, Solenne and Sylvie. | ||
| + | * Students: Eugene, Fabio, Hoang Vu, Juliette, Pierre, Romain, Sean and Terry. | ||
| + | {{Team:Paris_Saclay/notebook_footer}} | ||
Latest revision as of 14:24, 14 October 2014
Contents |
Monday 4th August
Lab Work
The chassis coli Odor free
Striate on Dishes
by Juliette & Romain
We screened MG1655 and MG1655Z1 colonies on LB and LB + Kanamycin dishes for the loss of kanamycin resistance cassette, using the colonies obtained from the streak plates prepared on LB dishes and incubated at 42°C. 1st August
Salicylate Inducible Suppressing System
Plasmid DNA Purification
by Eugene and Fabio
- Ligation of BBa_J61051 and BBa_K228001 made the 30th July
From Liquid Culture made the 1st August
Digestion to check
by Eugene and Fabio
In order to validate the hole Assembly Process, we did a final digestion. The samples were digested with the enzymes EcoRI and PstI, that separates the BioBrick Assembly core from theirs vectors. The graphic result of this process is shown in the next step.
Protocol:
- Add 5 μl of the plasmid to digest.
- Add 2 μl of buffer (Fast Digest Buffer 10x).
- Add 0,2 μl of enzyme EcoRI.
- Add 0,2 μl of enzyme PstI.
- Complete with 12,6μl of H2O.
- Mix gently.
- Incubate at 37°C for one hour.
Electrophoresis
by Fabio
The Electrophoresis was used to verify the success of the Digestion and the success of the plasmid DNA purification at the same time. Strain number 9 represents the digestion's control as it is the BBa_K228001, the BioBrick used as vector. If the clones have the same size of the control, it indicates that we had no ligation between both BioBricks (BBa_J61051 and BBa_K228001). Each sample has 20 μl and the Ladder has 5 μl.
- Plasmid Cl.1
- Plasmid Cl.2
- Plasmid Cl.3
- Plasmid Cl.4
- Plasmid Cl.5
- Plasmid Cl.6
- Plasmid Cl.7
- Plasmid Cl.8
- BBa_K228001 - Control
Results:
- Failed, the plasmid had no digestion.
- Success, the plasmid had the good size of digestion.
- Success, the plasmid had the good size of digestion.
- Success, the plasmid had the good size of digestion.
- Success, the plasmid had the good size of digestion.
- Success, the plasmid had the good size of digestion.
- Success, the plasmid had the good size of digestion.
- Failed, the plasmid had others digestions points.
- Good, we have a good control.
BioBrick Assembly - Segregate Process Protocol
Final Stock
by Fabio
As the BioBrick Assembly was a success, proved by the last Electrophoresis, we have registered a new BioBrick named BBa_K1372000 together with its final stock. We used the Clones 2, 3 and 5 from Liquid Colonies made the 1st August as the Electrophoresis shows a good concentration of them.
- BBa_K1372000 Cl. 1
- BBa_K1372000 Cl. 2
- BBa_K1372000 Cl. 3
1 ml of colonies plus 0.25 ml of glycerol 87%
D - Lemon scent
Gel electrophoresis of BBa_K762100
by Sean
Samples used were prepared on the 30th July.
Results
From left to right: 5 µl of BBa_K762100 + "old" Phusion enzyme, 5 µl of BBa_K762100 + "new" Phusion enzyme
Electroporation
by Terry
Strain used: DY330 containing plasmid pJBEI6409. We had to switch the Limonen Synthase cassette with the Apramycin resistance.
15 ml of culture has been placed at 42°C for 17 min then cooled 10 min before centrifugation.
Make 2 électroporations in cold electroporation cuvettes:
- A control cuvette: 50µl of DY330.
- A second cuvette: 50µl of DY330 culture + 8µl of PCR purified product identified the 21st July
Electroporation : 2500V, 132W, 40µF.
After that, add 1ml of cold LB in each cuvette and transfer in tubes to incubate during 1 to 2h at 30°C.
Spread on 4 dishes LB + Apramicine :
- 100µl of control
- 50µl of transformed DY330 with 8µl of PCR purified product
- 100µl of transformed DY330 with 8µl of PCR purified product
- The rest of transformed DY330 with 8µl of PCR purified product
Incubate for 2 days at 30°C.
Polymerase chain reaction
by Sean & Pierre
BBa_K517003
| component | volume |
|---|---|
| H2O | 35.5μl |
| Phusion buffer 5X | 10μl |
| dNTPs | 2μl |
| iPS68bis | 1μl |
| iPS69 | 1μl |
| DNA | 1μl |
| Phusion enzyme | 0.5μl |
BBa_K762100
For this BioBrick two tubes were prepared: one with undiluted DNA and one with DNA diluted 10-1.
| component | volume |
|---|---|
| H2O | 35.5μl |
| Phusion buffer 5X | 10μl |
| dNTPs | 2μl |
| iPS66 | 1μl |
| iPS67 | 1μl |
| DNA | 1μl |
| Phusion enzyme | 0.5μl |
Tubes were placed in PCR machine with the following parameters.
| Cycle step | Temperature | Time | Cycle |
|---|---|---|---|
|
Initial denaturation |
98°C |
1 min |
1 |
| Denaturation | 98°C | 15 s | 25 - 30 |
| Annealing | 52°C | 25 s | 25 - 30 |
| Extension | 72°C | 45 s | 25-30 |
| Final extension | 72°C | 10 min | 1 |
| Final extension | 8°C | hold | 1 |
Gel electrophoresis of BBa_K517003 and K762100 previously prepared
by Sean&Pierre
Results
failure.
From left to right: 10 µl ladder, 5 µl of BBa_K762100 non diluted, 5 µl of BBa_K762100 diluted 10-1, 5 µl of BBa_K517003.
Miscellaneous
By Hoang Vu
Ethics
Well, I just got back from one month of vacation and when I came, everybody was already on lab works. So, I decided to work on ethics first. Synthetic Biology and BioArt are controversial because one is about engineering living organisms, and therefore raises economic, religious, political, moral problems; and the other is about engineering the living organisms too but it's mostly USING the living organisms to do art, to carry a message. This is how I see the essential points of our project ethical problems. But my point of view is not fixed. Maybe I'm wrong, maybe there is something that I have missed. That's why I started with a research on GMOs in general to better understand those issues.
- The first trangenic pet were GloFish, "TK-1" 2003 in Taiwan
- California is completely against such animals but the funny thing is that California is one of the best supporters of agricultural GMOs. So, there is the question: Why the plants and not the fish? Probably economic benefits and ethical positions are the reasons. Then, is there a hierachy of living organisms? Why?
- GMOs under commercial cover raise new problems on environment and commercial rights.
- Take a look at the arguments between Anti-GMO and Pro-GMO movements. The 90's context for example in Europe after food crises like the mad cow disease, contribute to the incomplete acceptance by the Europeans.
Bibliography
I worked on ethics till one of our advisors ask me: "Can you do a research on the chemical components of the lemon odor please? I need more publications on chemical characterization of limonene, citral A and B, and beta-Pinene"
Photo of the Day
Members there:
- Instructors and advisors: Alice, Solenne and Sylvie.
- Students: Eugene, Fabio, Hoang Vu, Juliette, Pierre, Romain, Sean and Terry.
 "
"