Team:Paris Saclay/Notebook/July/29
From 2014.igem.org
(→PCR of BBa_K762100) |
m (→Tuesday 29th July) |
||
| (25 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{Team:Paris_Saclay/notebook_header}} | ||
=Tuesday 29th July= | =Tuesday 29th July= | ||
| - | |||
==Lab Work== | ==Lab Work== | ||
| - | + | ===The frame coli Odor free=== | |
| - | === | + | |
| - | + | ||
====PCR==== | ====PCR==== | ||
''by Romain'' | ''by Romain'' | ||
| - | + | Strains used: E. coli MG1655Z1 and E. coli MG1655, and oligonucleotides used: FTR-Apra-F and FTR-Apra-R. | |
| - | Protocol: | + | '''Protocol''' |
| + | |||
| + | Add into a PCR tube the following: | ||
| + | |||
| + | {| class="wikitable centre" width="50%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | Component | ||
| + | ! scope=col | For a total volume of 50μl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |27.25μl | ||
| + | |- | ||
| + | | Green GoTaq buffer 5X | ||
| + | | 10μl | ||
| + | |- | ||
| + | | dNTPs | ||
| + | | 1μl | ||
| + | |- | ||
| + | | FTR-Apra-F | ||
| + | | 2μl | ||
| + | |- | ||
| + | | FTR-Apra-R | ||
| + | | 2μl | ||
| + | |- | ||
| + | | DMSO | ||
| + | | 1.5μl | ||
| + | |- | ||
| + | | MgCl<sub>2</sub> | ||
| + | | 4μl | ||
| + | |- | ||
| + | | Bacterial culture | ||
| + | | 2μl | ||
| + | |- | ||
| + | | Green GoTaq enzyme | ||
| + | | 0.25μl | ||
| + | |} | ||
| + | |||
| + | Tube was placed in PCR machine with the following parameters. | ||
| + | |||
| + | {| class="wikitable centre" width="80%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | Cycle step | ||
| + | ! scope=col | Temperature | ||
| + | ! scope=col | Time | ||
| + | ! scope=col | Cycle | ||
| + | |- | ||
| + | | width="25%" | | ||
| + | Initial denaturation | ||
| + | | width="25%" | | ||
| + | 95°C | ||
| + | | width="25%" | | ||
| + | 5 min | ||
| + | | width="25%" | | ||
| + | 1 | ||
| + | |- | ||
| + | | Denaturation | ||
| + | | 95°C | ||
| + | | 30 s | ||
| + | | 30 | ||
| + | |- | ||
| + | | Annealing | ||
| + | | 60°C | ||
| + | | 30 s | ||
| + | | 30 | ||
| + | |- | ||
| + | | Extension | ||
| + | | 72°C | ||
| + | | 2 min | ||
| + | | 30 | ||
| + | |- | ||
| + | | Final extension | ||
| + | | 72°C | ||
| + | | 5 min | ||
| + | | 1 | ||
| + | |- | ||
| + | | Final extension | ||
| + | | 12°C | ||
| + | | hold | ||
| + | | 1 | ||
| + | |} | ||
| - | + | The different bacterial cultures in each tubes: | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
#Tube 1: MG1655Z1 10I | #Tube 1: MG1655Z1 10I | ||
| Line 30: | Line 101: | ||
#Tube 7: MG1655Z1 10I positive control with plasmid | #Tube 7: MG1655Z1 10I positive control with plasmid | ||
| - | + | Results of the PCR: The electrophoresis revealed nothing | |
| - | + | ||
| - | + | New PCR with different enyme. | |
| - | + | ||
| - | + | ||
| - | === | + | Strains used: MG1655 and MG1655Z1, and oligonucleotides used: FTR-Apra-F and FTR-Apra-R. |
| + | |||
| + | '''Protocol''' | ||
| + | |||
| + | Add into a PCR tube the following: | ||
| + | |||
| + | {| class="wikitable centre" width="50%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | Component | ||
| + | ! scope=col | For a total volume of 50μl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |37.25μl | ||
| + | |- | ||
| + | | DreamTaq buffer 10X | ||
| + | | 5μl | ||
| + | |- | ||
| + | | dNTPs | ||
| + | | 1μl | ||
| + | |- | ||
| + | | FTR-Apra-F | ||
| + | | 1μl | ||
| + | |- | ||
| + | | FTR-Apra-R | ||
| + | | 1μl | ||
| + | |- | ||
| + | | DMSO | ||
| + | | 2.5μl | ||
| + | |- | ||
| + | | Bacterial culture | ||
| + | | 2μl | ||
| + | |- | ||
| + | | DreamTaq enzyme | ||
| + | | 0.25μl | ||
| + | |} | ||
| + | |||
| + | Tube was placed in PCR machine with the following parameters. | ||
| + | |||
| + | {| class="wikitable centre" width="80%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | Cycle step | ||
| + | ! scope=col | Temperature | ||
| + | ! scope=col | Time | ||
| + | ! scope=col | Cycle | ||
| + | |- | ||
| + | | width="25%" | | ||
| + | Initial denaturation | ||
| + | | width="25%" | | ||
| + | 95°C | ||
| + | | width="25%" | | ||
| + | 5 min | ||
| + | | width="25%" | | ||
| + | 1 | ||
| + | |- | ||
| + | | Denaturation | ||
| + | | 95°C | ||
| + | | 30 s | ||
| + | | 30 | ||
| + | |- | ||
| + | | Annealing | ||
| + | | 60°C | ||
| + | | 30 s | ||
| + | | 30 | ||
| + | |- | ||
| + | | Extension | ||
| + | | 72°C | ||
| + | | 2 min | ||
| + | | 30 | ||
| + | |- | ||
| + | | Final extension | ||
| + | | 72°C | ||
| + | | 5 min | ||
| + | | 1 | ||
| + | |- | ||
| + | | Final extension | ||
| + | | 12°C | ||
| + | | hold | ||
| + | | 1 | ||
| + | |} | ||
| + | |||
| + | #Tube 1: MG1655Z1 10I | ||
| + | #Tube 2: MG1655 100II | ||
| + | #Tube 3: MG1655 50I | ||
| + | #Tube 4: MG1655 100I | ||
| + | #Tube 5: MG1655Z1 10II | ||
| + | #Tube 6: MG1655 50II | ||
| + | #Tube 7: MG1655Z1 10I positive control with plasmid | ||
| + | |||
| + | Results of the PCR: The electrophoresis revealed successfully the DNA! | ||
| + | |||
| + | ===Salicylate Inducible Suppressing System=== | ||
| + | ====Digestion==== | ||
| + | ''by Fabio'' | ||
| + | |||
| + | After the experiments done [https://2014.igem.org/Team:Paris_Saclay/Notebook/July/28#C_-_Salicylate_Inducible_Suppressing_System yesterday], we are sure to manipulate the right plasmid from both BioBricks and also that we have a good concentration of them. Now it's time to start the [http://parts.igem.org/Help:Assembly/3A_Assembly Assembly procedure] in order to concatenate both BioBricks. | ||
| + | |||
| + | * BioBrick '''BBa_J61051''' (Salicylate promoter + NahR) as '''Part A''' | ||
| + | * BioBrick '''BBa_K228001''' (RNA suppressor) as '''Part B''' | ||
| + | |||
| + | The enzymes used were: | ||
| + | |||
| + | * '''EcoRI''' and '''SpeI''' for '''BBa_J61051''' | ||
| + | * '''PstI''' and '''XbaI''' for '''BBa_K228001''' | ||
| + | |||
| + | <font color="#F00">TODO: Illustration of the process</font> | ||
| + | |||
| + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/BioBrick_Assembly#Digest_reaction BioBrick Assembly - Digest Reaction Protocol] | ||
| + | |||
| + | ====Segregate Process by Electrophoresis==== | ||
| + | [[File:LU000107a.jpg|right|300px]] | ||
| + | [[File:RL007744.jpg|right|300px]] | ||
| + | ''by Fabio'' | ||
| + | |||
| + | First we tested both products of the digestions in order to verify the success of the reaction (''Process A''), which remained for 25 minutes in 100V. Once the sizes of the digestion's products confirmed, we did the ''Process B'' (1 hour in 100V) in order to segregate a good amount of DNA to the next step. | ||
| + | |||
| + | '''''Process A''''' | ||
| + | # BBa_J61051 | ||
| + | # BBa_K228001 | ||
| + | |||
| + | '''''Process B''''' | ||
| + | # BBa_J61051 ''strains from the bottom band of '''process A''''' | ||
| + | |||
| + | '''Results:''' | ||
| + | *'''A''' #1: Success, we can clearly see the vector and the core of the BioBrick after the digestion. | ||
| + | *'''A''' #2: Success, the digested BioBrick was cut one time and has the expected size. | ||
| + | *'''B''': Success, we have a very good concentration of the BBa_J61051's core. | ||
| + | |||
| + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/BioBrick_Assembly#Segregate_process BioBrick Assembly - Segregate Process Protocol] | ||
| + | |||
| + | ===Lemon scent=== | ||
| + | ====Transformation of electrocompetent cells==== | ||
| + | ''by Arnaud & Terry'' | ||
| + | |||
| + | Strain used: DY330. Plasmid used: pJBEI-6409 (code for enzymes to produce limonene from glucose in E. coli) | ||
| + | |||
| + | Make 2 électroporations in cold electroporation cuvettes: | ||
| + | |||
| + | *A control cuvette(without DNA): 50µl of DY330. | ||
| + | *A second cuvette: 50µl of DY330 culture + 1µl of pJBEI-6409 plasmid. | ||
| + | |||
| + | Electroporation : 2500V, 132W, 40µF. | ||
| + | |||
| + | After that, add 1ml of cold LB in each cuvette and transfer in 2 tubes to incubate during 1 to 2h at 30°C. | ||
| + | |||
| + | Spread on 4 dishes LB + Cm: | ||
| + | |||
| + | *100µl of control (without plasmid) | ||
| + | *50µl of transformed DY330 with pJBEI-6409 | ||
| + | *100µl of transformed DY330 with pJBEI-6409 | ||
| + | *The rest of transformed DY330 with pJBEI-6409 (concentrate in approximately 150µl) | ||
| + | |||
| + | Incubate for the weekend at 30°C. | ||
====PCR of BBa_K762100==== | ====PCR of BBa_K762100==== | ||
| Line 78: | Line 299: | ||
|} | |} | ||
| - | [ | + | Tube was placed in PCR machine with the following parameters. |
| + | |||
| + | {| class="wikitable centre" width="80%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | Cycle step | ||
| + | ! scope=col | Temperature | ||
| + | ! scope=col | Time | ||
| + | ! scope=col | Cycle | ||
| + | |- | ||
| + | | width="25%" | | ||
| + | Initial denaturation | ||
| + | | width="25%" | | ||
| + | 98°C | ||
| + | | width="25%" | | ||
| + | 1 min | ||
| + | | width="25%" | | ||
| + | 1 | ||
| + | |- | ||
| + | | Denaturation | ||
| + | | 98°C | ||
| + | | 15 s | ||
| + | | 25 - 30 | ||
| + | |- | ||
| + | | Annealing | ||
| + | | 55°C | ||
| + | | variable | ||
| + | | 25 - 30 | ||
| + | |- | ||
| + | | Extension | ||
| + | | 72°C | ||
| + | | 45 s | ||
| + | | 25-30 | ||
| + | |- | ||
| + | | Final extension | ||
| + | | 72°C | ||
| + | | 10 min | ||
| + | | 1 | ||
| + | |- | ||
| + | | Final extension | ||
| + | | 8°C | ||
| + | | hold | ||
| + | | 1 | ||
| + | |} | ||
| + | |||
| + | |||
| + | ==Photo of the Day== | ||
| + | [[File:Paris Saclay 29_july.jpg|600px|center]] | ||
| + | |||
| + | '''Members there''': | ||
| + | * Instructors and advisors: Alice, Solenne and Sylvie. | ||
| + | * Students: Arnaud, Fabio, Romain, Sean and Terry. | ||
| + | {{Team:Paris_Saclay/notebook_footer}} | ||
Latest revision as of 15:44, 14 October 2014
Contents |
Tuesday 29th July
Lab Work
The frame coli Odor free
PCR
by Romain
Strains used: E. coli MG1655Z1 and E. coli MG1655, and oligonucleotides used: FTR-Apra-F and FTR-Apra-R.
Protocol
Add into a PCR tube the following:
| Component | For a total volume of 50μl |
|---|---|
| H2O | 27.25μl |
| Green GoTaq buffer 5X | 10μl |
| dNTPs | 1μl |
| FTR-Apra-F | 2μl |
| FTR-Apra-R | 2μl |
| DMSO | 1.5μl |
| MgCl2 | 4μl |
| Bacterial culture | 2μl |
| Green GoTaq enzyme | 0.25μl |
Tube was placed in PCR machine with the following parameters.
| Cycle step | Temperature | Time | Cycle |
|---|---|---|---|
|
Initial denaturation |
95°C |
5 min |
1 |
| Denaturation | 95°C | 30 s | 30 |
| Annealing | 60°C | 30 s | 30 |
| Extension | 72°C | 2 min | 30 |
| Final extension | 72°C | 5 min | 1 |
| Final extension | 12°C | hold | 1 |
The different bacterial cultures in each tubes:
- Tube 1: MG1655Z1 10I
- Tube 2: MG1655 100II
- Tube 3: MG1655 50I
- Tube 4: MG1655 100I
- Tube 5: MG1655Z1 10II
- Tube 6: MG1655 50II
- Tube 7: MG1655Z1 10I positive control with plasmid
Results of the PCR: The electrophoresis revealed nothing
New PCR with different enyme.
Strains used: MG1655 and MG1655Z1, and oligonucleotides used: FTR-Apra-F and FTR-Apra-R.
Protocol
Add into a PCR tube the following:
| Component | For a total volume of 50μl |
|---|---|
| H2O | 37.25μl |
| DreamTaq buffer 10X | 5μl |
| dNTPs | 1μl |
| FTR-Apra-F | 1μl |
| FTR-Apra-R | 1μl |
| DMSO | 2.5μl |
| Bacterial culture | 2μl |
| DreamTaq enzyme | 0.25μl |
Tube was placed in PCR machine with the following parameters.
| Cycle step | Temperature | Time | Cycle |
|---|---|---|---|
|
Initial denaturation |
95°C |
5 min |
1 |
| Denaturation | 95°C | 30 s | 30 |
| Annealing | 60°C | 30 s | 30 |
| Extension | 72°C | 2 min | 30 |
| Final extension | 72°C | 5 min | 1 |
| Final extension | 12°C | hold | 1 |
- Tube 1: MG1655Z1 10I
- Tube 2: MG1655 100II
- Tube 3: MG1655 50I
- Tube 4: MG1655 100I
- Tube 5: MG1655Z1 10II
- Tube 6: MG1655 50II
- Tube 7: MG1655Z1 10I positive control with plasmid
Results of the PCR: The electrophoresis revealed successfully the DNA!
Salicylate Inducible Suppressing System
Digestion
by Fabio
After the experiments done yesterday, we are sure to manipulate the right plasmid from both BioBricks and also that we have a good concentration of them. Now it's time to start the [http://parts.igem.org/Help:Assembly/3A_Assembly Assembly procedure] in order to concatenate both BioBricks.
- BioBrick BBa_J61051 (Salicylate promoter + NahR) as Part A
- BioBrick BBa_K228001 (RNA suppressor) as Part B
The enzymes used were:
- EcoRI and SpeI for BBa_J61051
- PstI and XbaI for BBa_K228001
TODO: Illustration of the process
BioBrick Assembly - Digest Reaction Protocol
Segregate Process by Electrophoresis
by Fabio
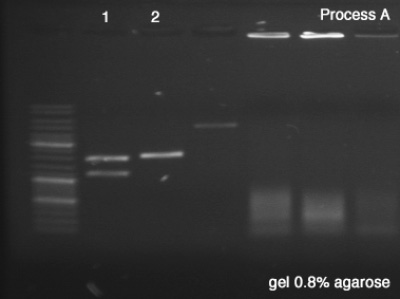
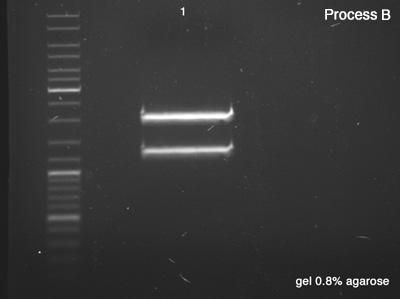
First we tested both products of the digestions in order to verify the success of the reaction (Process A), which remained for 25 minutes in 100V. Once the sizes of the digestion's products confirmed, we did the Process B (1 hour in 100V) in order to segregate a good amount of DNA to the next step.
Process A
- BBa_J61051
- BBa_K228001
Process B
- BBa_J61051 strains from the bottom band of process A
Results:
- A #1: Success, we can clearly see the vector and the core of the BioBrick after the digestion.
- A #2: Success, the digested BioBrick was cut one time and has the expected size.
- B: Success, we have a very good concentration of the BBa_J61051's core.
BioBrick Assembly - Segregate Process Protocol
Lemon scent
Transformation of electrocompetent cells
by Arnaud & Terry
Strain used: DY330. Plasmid used: pJBEI-6409 (code for enzymes to produce limonene from glucose in E. coli)
Make 2 électroporations in cold electroporation cuvettes:
- A control cuvette(without DNA): 50µl of DY330.
- A second cuvette: 50µl of DY330 culture + 1µl of pJBEI-6409 plasmid.
Electroporation : 2500V, 132W, 40µF.
After that, add 1ml of cold LB in each cuvette and transfer in 2 tubes to incubate during 1 to 2h at 30°C.
Spread on 4 dishes LB + Cm:
- 100µl of control (without plasmid)
- 50µl of transformed DY330 with pJBEI-6409
- 100µl of transformed DY330 with pJBEI-6409
- The rest of transformed DY330 with pJBEI-6409 (concentrate in approximately 150µl)
Incubate for the weekend at 30°C.
PCR of BBa_K762100
by Sean
Oligonucleotides used: iPS66, iPS67
Oligonucleotides were diluted twice from 100µM to 50µM.
Protocol
Add into a PCR tube the following:
| Component | For a total volume of 50μl |
|---|---|
| H2O | 35.5μl |
| Phusion buffer 5X | 10μl |
| dNTPs | 1μl |
| iPS66 | 1μl |
| iPS67 | 1μl |
| BBa_K762100 | 1μl |
| Phusion DNA polymerase | 0.25μl |
Tube was placed in PCR machine with the following parameters.
| Cycle step | Temperature | Time | Cycle |
|---|---|---|---|
|
Initial denaturation |
98°C |
1 min |
1 |
| Denaturation | 98°C | 15 s | 25 - 30 |
| Annealing | 55°C | variable | 25 - 30 |
| Extension | 72°C | 45 s | 25-30 |
| Final extension | 72°C | 10 min | 1 |
| Final extension | 8°C | hold | 1 |
Photo of the Day
Members there:
- Instructors and advisors: Alice, Solenne and Sylvie.
- Students: Arnaud, Fabio, Romain, Sean and Terry.
 "
"