Team:Paris Saclay/Notebook/August/12
From 2014.igem.org
Terry.Mara (Talk | contribs) (→Human Pratices) |
(→Lab work) |
||
| Line 2: | Line 2: | ||
=Tuesday 12th August= | =Tuesday 12th August= | ||
==Lab work== | ==Lab work== | ||
| + | ===A - The chassis coli Odor free=== | ||
| + | ====Competents cells (CaCl2)==== | ||
| + | ''by Melanie'' | ||
| + | |||
| + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/Transformation_of_competent_e.coli_by_CaCl2#Transformation protocol] | ||
| + | |||
| + | stock of competents cells (odor free) | ||
| + | |||
| + | ===pJBEI transformation=== | ||
| + | |||
| + | we use odor free e.coli | ||
| + | |||
===C - Salicylate Inducible Suppressing System=== | ===C - Salicylate Inducible Suppressing System=== | ||
====Plasmid DNA Purification==== | ====Plasmid DNA Purification==== | ||
| Line 105: | Line 117: | ||
|0.5μl | |0.5μl | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
==Human Pratices== | ==Human Pratices== | ||
Revision as of 07:54, 19 August 2014
Contents |
Tuesday 12th August
Lab work
A - The chassis coli Odor free
Competents cells (CaCl2)
by Melanie
stock of competents cells (odor free)
pJBEI transformation
we use odor free e.coli
C - Salicylate Inducible Suppressing System
Plasmid DNA Purification
by Fabio
- BBa_K1372001 Cl.1
- BBa_K1372001 Cl.2
From Liquid Culture made the 11th August
Digestion to check
by Fabio
In order to validate the hole Assembly Process, we did a final digestion. The samples were digested with the enzymes EcoRI and PstI, that separates the BioBrick Assembly core from theirs vectors. The graphic result of this process is shown in the next step.
Protocol:
- Add 5 μl of the plasmid to digest.
- Add 2 μl of buffer (Fast Digest Buffer 10x).
- Add 0,2 μl of enzyme EcoRI.
- Add 0,2 μl of enzyme PstI.
- Complete with 12,6μl of H2O.
- Mix gently.
- Incubate at 37°C for one hour.
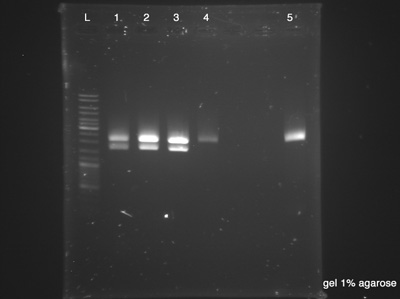
Electrophoresis
by Fabio
The Electrophoresis was used to verify the success of the Digestion and the success of the plasmid DNA purification at the same time. Strain number 5 represents the digestion's control as it is the BBa_B0015, the BioBrick used as vector. If the clones have the same size of the control, it indicates that we had no ligation between both BioBricks (BBa_K1372000 and BBa_B0015). Each sample has 20 μl and the Ladder has 10 μl.
- Plasmid Cl.1
- Plasmid Cl.2
- Plasmid Cl.3
- Plasmid Cl.4
- BBa_B0015 - Control
Results:
- Success, vector has 2200 bp and insert has 1533 bp.
- Success, vector has 2200 bp and insert has 1533 bp.
- Success, vector has 2200 bp and insert has 1533 bp.
- Failed, the plasmid had no digestion.
- Good, we have a control.
BioBrick Assembly - Segregate Process Protocol
Final Stock
by Fabio
As the BioBrick Assembly was a success, proved by the last Electrophoresis, we have registered a new BioBrick named BBa_K1372001 together with its final stock. We used the Clones 2 and 3 from Liquid Colonies made the 11th August as the Electrophoresis shows a good concentration of them.
- BBa_K1372001 Cl. 1
- BBa_K1372001 Cl. 2
1 ml of colonies plus 0.20 ml of glycerol 87%
D - Lemon scent
PCR Clean-up of p cola and BBa_K517003
by Sean
PCR prepared on the 7th August.
Clean-up performed with the following protocol.
NB: in the present case we have 40 µl of each PCR result and we use 20 µl of elution buffer.
Gel electrophoresis of p cola, BBa_K762100 and BBa_K517003
by Sean
Samples used: PCR clean-up results from 8th August (1 µl of each).
Results: BBa_K762100 has barely migrated. Since this was prepared with Phusion, we will perform a PCR with GoTaq (as was the case for p cola and BBa_K517003)
PCR of BBa_K762100
Five tubes were prepared. The following is for one tube.
| component | volume |
|---|---|
| H2O | 27.25μl |
| GoTaq buffer 5X | 10μl |
| MgCl2 | 4μl |
| DMSO | 1.5μl |
| dNTPs | 2μl |
| iPS66 (10μM) | 1μl |
| iPS67 (10μM) | 1μl |
| DNA | 1μl |
| GoTaq enzyme | 0.5μl |
Human Pratices
Art & Design
by Terry
Resumption of the preculture made yesterday, expressing blue chromoprotein. We will make 4 pieces of agar ( + control ) with different concentrations of bacteria in it. The chromoprotein is expressed in presence of dioxygene. The liquide preculture was not blue because the incubation has been made in a closed Falcon.
Protocol:
- Preparation of a stock of 200ml agar gel at 20mg/ml. ( 4g of solid agar in 200ml of hot water )
- Sterilisation of the mold with Ethanol 70%, all the work will be made in steril condition.
- Control of the Optic Density : 0.86
- 4 dilutions are prepared in total volume of 1ml :
- 1ml of preculture (100%)
- 500µl of LB and 500µl of preculture (50%)
- 900µl of LB and 100µl of preculture (10%)
- 990µl of LB and 10µl of preculture (1%)
- Introduction of 1ml of the different dilutions in the molds
- Introduction of 10ml of semi-liquid agar in each mold. Agar's temperature : 40°C
- The molds are securised with Aluminium.
- Direct incubation at 37°C
 "
"