Team:Paris Saclay/Notebook/July/24
From 2014.igem.org
(→Thursday 24th July) |
m (→Plasmid DNA Extraction and Electrophoresis) |
||
| (37 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{Team:Paris_Saclay/notebook_header}} | ||
=Thursday 24th July= | =Thursday 24th July= | ||
| - | |||
==Lab work== | ==Lab work== | ||
| + | ===Construction of the fusion protein (color)=== | ||
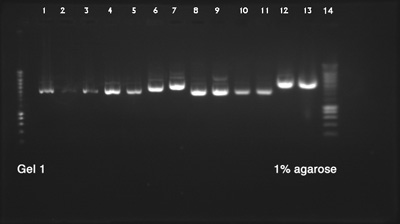
| + | ====Plasmid DNA Extraction and Electrophoresis==== | ||
| + | ''by Terry'' | ||
| - | + | [[File:Electrophoresis_24.07.14.jpg|400px|right]] | |
| - | + | Biobricks used: | |
| + | # BBa_K1033927 with pSB1C3 (pink chromoprotein) | ||
| + | # BBa_K1033910 with pSB1C3 (yellow chromoprotein) | ||
| + | # BBa_K1033921 with pSB1C3 (red chromoprotein) | ||
| + | # BBa_K1033922 with pSB1C3 (red chromoprotein) | ||
| + | # BBa_K1033902 with pSB1C3 (blue chromoprotein) | ||
| + | # BBa_K731201 with pSB1C3 (Arabinose inductible araC-pBAD promoter) | ||
| + | # BBa_K1033925 with pSB1C3 (pink chromoprotein) | ||
| + | # BBa_K1033905 with pSB1C3 (purple chromoprotein) | ||
| + | # BBa_K1033913 with pSB1C3 (orange chromoprotein) | ||
| + | # BBa_K1033929 with pSB1C3 (blue chromoprotein) | ||
| + | '''Result:''' | ||
| + | |||
| + | Success, all DNAs have the expected size | ||
| + | |||
| + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/Extraction_of_the_Genomic_DNA_from_Bacteria_by_using_NucleoSpin%C2%AE_Tissue Protocol] | ||
| + | |||
| + | === Salicylate Inducible Suppressing System=== | ||
| + | ====Plasmid DNA Extraction & Electrophoresis==== | ||
| + | ''by Terry'' | ||
| + | |||
| + | Biobricks used: | ||
| + | *12 BBa_J45017 with pSB1C3 | ||
| + | *13 BBa_J45017 with pSB1AK3 (pchBA synthase (converts chorismate to salicylate)) | ||
| + | |||
| + | (12 and 13 = on the previous gel) | ||
| + | |||
| + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/Extraction_of_the_Genomic_DNA_from_Bacteria_by_using_NucleoSpin%C2%AE_Tissue Protocol] | ||
| + | |||
| + | ====Transformation of competent E.coli cells==== | ||
| + | ''by Fabio'' | ||
| + | |||
| + | We transformed in DH5α both BioBricks rehydrated [https://2014.igem.org/Team:Paris_Saclay/Notebook/July/23 yesterday] ('''BBa_J61051''' and '''BBa_K228001'''). | ||
| + | |||
| + | '''Samples:''' | ||
| + | # BBa_J61051 Concentrated | ||
| + | # BBa_J61051 Undiluted | ||
| + | # BBa_K228001 Concentrated | ||
| + | # BBa_K228001 Undiluted | ||
| + | # DH5α Control | ||
| + | |||
| + | '''Results on 25th July morning:''' | ||
| + | # Success, grown very well (> 200 colonies) | ||
| + | # Success, grown well (> 50 colonies) | ||
| + | # Success, grown well (> 90 colonies) | ||
| + | # Success, grown (15 colonies) | ||
| + | # Success, nothing has grown | ||
| + | |||
| + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/Transformation_of_supercompetent_Ecoli_cells_with_CaCl2 Protocol] | ||
| + | |||
| + | ===Lemon scent=== | ||
| + | ====Plasmid DNA Extraction & Electrophoresis==== | ||
| + | |||
| + | ''by Terry'' | ||
| + | |||
| + | Biobricks used: | ||
| + | *7. BBa_K762100 with pSB1C3 (Limonen synthase1 with RBS) | ||
| + | |||
| + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/Extraction_of_the_Genomic_DNA_from_Bacteria_by_using_NucleoSpin%C2%AE_Tissue Protocol] | ||
| + | |||
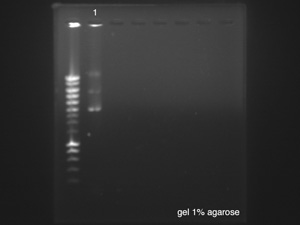
| + | ====Gel electrophoresis of p cola==== | ||
| + | ''by Sean'' | ||
| + | |||
| + | [[File:LU000081.jpg|right]] | ||
| + | |||
| + | p cola used: sample left overnight [https://2014.igem.org/Team:Paris_Saclay/Notebook/July/23 yesterday] resuspended in 30μl of H<sub>2</sub>O milliQ. | ||
| + | |||
| + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/Gel_Electrophoresis Protocol] | ||
| + | |||
| + | ====Results of the PCR Targeting==== | ||
''by Romain'' | ''by Romain'' | ||
PCR targeting made the [https://2014.igem.org/Team:Paris_Saclay/Notebook/July/22 July 22nd.] | PCR targeting made the [https://2014.igem.org/Team:Paris_Saclay/Notebook/July/22 July 22nd.] | ||
| + | Strains used: DY330 (clone I and II). Plasmid used: pJBEI-6409 (code for enzymes to produce limonene from glucose in E. coli) | ||
Results: | Results: | ||
| - | |||
*Nothing has grown on the control dishes. | *Nothing has grown on the control dishes. | ||
*Failure, nothing has grown on the clone I dishes. | *Failure, nothing has grown on the clone I dishes. | ||
*Failure, other bacteria have grown on the clone II dishes. | *Failure, other bacteria have grown on the clone II dishes. | ||
| - | We have to make another PCR. | + | We have to make another PCR. Liquid culture of DY330 made in 30ml LB. |
| + | |||
| + | New PCR of the oligonucleotides iPS70 and iPS71 made with the enzyme DreamTaq. | ||
| + | |||
| + | Protocol: | ||
| + | *108,75µl of H<sub>2</sub>O MilliQ | ||
| + | *15µl DreamTaq buffer | ||
| + | *6µl oligonucleotide iPS70 | ||
| + | *6µl oligonucleotide iPS71 | ||
| + | *4,5µl DMSO | ||
| + | *6µl Plasmid Posv230 diluted | ||
| + | *3µl dNTP | ||
| + | *0,75µl of DreamTaq enzyme | ||
| + | |||
| + | Results: After the PCR, the electrophoresis has revealed nothing. | ||
| + | |||
| + | So we made another PCR of the oligonucleotides iPS70 and iPS71 with the GoTaq enzyme. | ||
| + | |||
| + | *56,5µl of H2O | ||
| + | *20µl of GoTaq Flexi Green buffer (5X) | ||
| + | *4µl oligonucleotide iPS70 | ||
| + | *4µl oligonucleotide iPS71 | ||
| + | *3µl DMSO | ||
| + | *2µl Plasmid Posv 230 no dilute | ||
| + | *2µl dNTP | ||
| + | *8µl MgCl2 | ||
| + | *0,5µl GoTaq Green enzyme | ||
| + | |||
| + | PCR program: 95°C during 30seconds, then [95°C during 30sec + 60°C during 30sec + 72°C during 2min] (30 cycles) then 72°C during 5min, and then 12°C to infinity. | ||
| + | |||
| + | Results [https://2014.igem.org/Team:Paris_Saclay/Notebook/July/25 tomorrow] | ||
| + | |||
| + | |||
| + | ==Photo of the Day== | ||
| + | [[File:Paris Saclay 24_july.jpg|600px|center]] | ||
'''Members present:''' | '''Members present:''' | ||
* Instructors and advisors: Alice and Solenne. | * Instructors and advisors: Alice and Solenne. | ||
* Students: Arnaud, Fabio, Juliette, Pierre, Romain, Sean and Terry. | * Students: Arnaud, Fabio, Juliette, Pierre, Romain, Sean and Terry. | ||
| - | + | {{Team:Paris_Saclay/notebook_footer}} | |
| - | + | ||
Latest revision as of 14:23, 17 October 2014
Contents |
Thursday 24th July
Lab work
Construction of the fusion protein (color)
Plasmid DNA Extraction and Electrophoresis
by Terry
Biobricks used:
- BBa_K1033927 with pSB1C3 (pink chromoprotein)
- BBa_K1033910 with pSB1C3 (yellow chromoprotein)
- BBa_K1033921 with pSB1C3 (red chromoprotein)
- BBa_K1033922 with pSB1C3 (red chromoprotein)
- BBa_K1033902 with pSB1C3 (blue chromoprotein)
- BBa_K731201 with pSB1C3 (Arabinose inductible araC-pBAD promoter)
- BBa_K1033925 with pSB1C3 (pink chromoprotein)
- BBa_K1033905 with pSB1C3 (purple chromoprotein)
- BBa_K1033913 with pSB1C3 (orange chromoprotein)
- BBa_K1033929 with pSB1C3 (blue chromoprotein)
Result:
Success, all DNAs have the expected size
Salicylate Inducible Suppressing System
Plasmid DNA Extraction & Electrophoresis
by Terry
Biobricks used:
- 12 BBa_J45017 with pSB1C3
- 13 BBa_J45017 with pSB1AK3 (pchBA synthase (converts chorismate to salicylate))
(12 and 13 = on the previous gel)
Transformation of competent E.coli cells
by Fabio
We transformed in DH5α both BioBricks rehydrated yesterday (BBa_J61051 and BBa_K228001).
Samples:
- BBa_J61051 Concentrated
- BBa_J61051 Undiluted
- BBa_K228001 Concentrated
- BBa_K228001 Undiluted
- DH5α Control
Results on 25th July morning:
- Success, grown very well (> 200 colonies)
- Success, grown well (> 50 colonies)
- Success, grown well (> 90 colonies)
- Success, grown (15 colonies)
- Success, nothing has grown
Lemon scent
Plasmid DNA Extraction & Electrophoresis
by Terry
Biobricks used:
- 7. BBa_K762100 with pSB1C3 (Limonen synthase1 with RBS)
Gel electrophoresis of p cola
by Sean
p cola used: sample left overnight yesterday resuspended in 30μl of H2O milliQ.
Results of the PCR Targeting
by Romain
PCR targeting made the July 22nd. Strains used: DY330 (clone I and II). Plasmid used: pJBEI-6409 (code for enzymes to produce limonene from glucose in E. coli)
Results:
- Nothing has grown on the control dishes.
- Failure, nothing has grown on the clone I dishes.
- Failure, other bacteria have grown on the clone II dishes.
We have to make another PCR. Liquid culture of DY330 made in 30ml LB.
New PCR of the oligonucleotides iPS70 and iPS71 made with the enzyme DreamTaq.
Protocol:
- 108,75µl of H2O MilliQ
- 15µl DreamTaq buffer
- 6µl oligonucleotide iPS70
- 6µl oligonucleotide iPS71
- 4,5µl DMSO
- 6µl Plasmid Posv230 diluted
- 3µl dNTP
- 0,75µl of DreamTaq enzyme
Results: After the PCR, the electrophoresis has revealed nothing.
So we made another PCR of the oligonucleotides iPS70 and iPS71 with the GoTaq enzyme.
- 56,5µl of H2O
- 20µl of GoTaq Flexi Green buffer (5X)
- 4µl oligonucleotide iPS70
- 4µl oligonucleotide iPS71
- 3µl DMSO
- 2µl Plasmid Posv 230 no dilute
- 2µl dNTP
- 8µl MgCl2
- 0,5µl GoTaq Green enzyme
PCR program: 95°C during 30seconds, then [95°C during 30sec + 60°C during 30sec + 72°C during 2min] (30 cycles) then 72°C during 5min, and then 12°C to infinity.
Results tomorrow
Photo of the Day
Members present:
- Instructors and advisors: Alice and Solenne.
- Students: Arnaud, Fabio, Juliette, Pierre, Romain, Sean and Terry.
 "
"