Team:Paris Saclay/Modeling/Fusion Proteine
From 2014.igem.org
m (→The second part – blue chromoprotein :) |
m (→The first part – Yellow chromoprotein:) |
||
| (19 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Paris_Saclay/modeling_header}} | {{Team:Paris_Saclay/modeling_header}} | ||
=Fusion Protein= | =Fusion Protein= | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Here we will discuss the structural modeling of the fusion protein that we had planned in order to obtain a [https://2014.igem.org/Team:Paris_Saclay/Project/Salicylate_Inducible_System green chromoprotein]. We used the bioinformatics tools developed by the Swiss Institute of Bioinformatics (Swiss-Model '''[1]''') and by the Xu group of Chicago University (RaptorX '''[2]'''). | Here we will discuss the structural modeling of the fusion protein that we had planned in order to obtain a [https://2014.igem.org/Team:Paris_Saclay/Project/Salicylate_Inducible_System green chromoprotein]. We used the bioinformatics tools developed by the Swiss Institute of Bioinformatics (Swiss-Model '''[1]''') and by the Xu group of Chicago University (RaptorX '''[2]'''). | ||
| Line 15: | Line 6: | ||
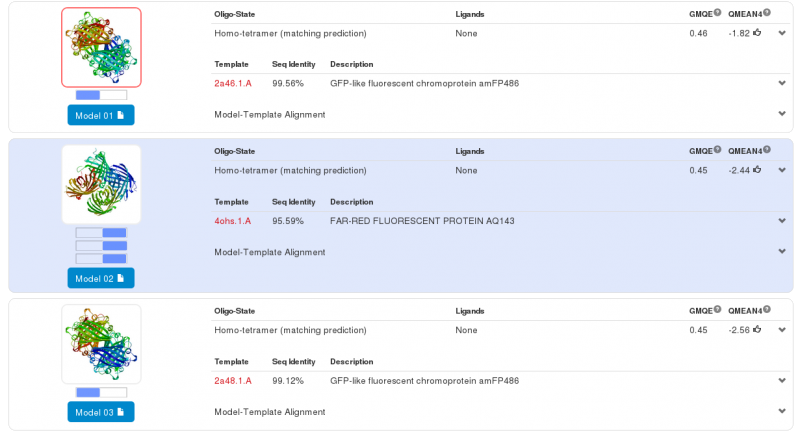
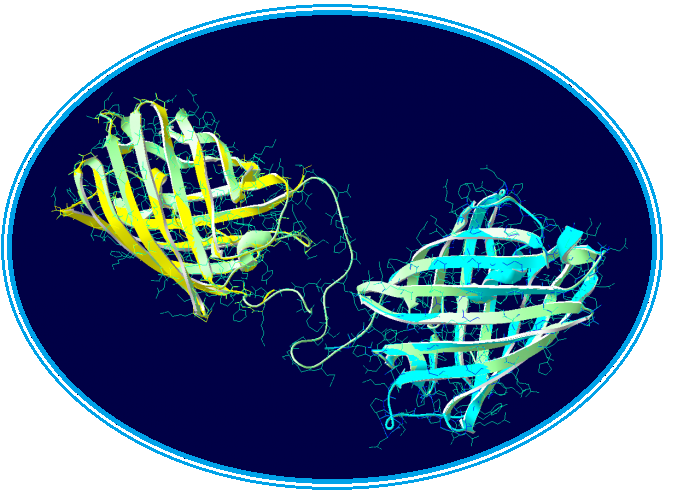
Swiss-Model has given us three highly probable models. Two models for the first part of our Fusion Protein and another for second part of our protein. We chose the first model for the yellow chromoprotein (1st part) and the second model for the blue chromoprotein (2nde part). | Swiss-Model has given us three highly probable models. Two models for the first part of our Fusion Protein and another for second part of our protein. We chose the first model for the yellow chromoprotein (1st part) and the second model for the blue chromoprotein (2nde part). | ||
| - | [[File:Paris_Saclay_SwissProt1.png|center| | + | [[File:Paris_Saclay_SwissProt1.png|center|660px]] |
| + | |||
| - | ==The first part – Yellow | + | ==The first part – Yellow chromoprotein:== |
| - | The yellow chromoprotein used to create this fusion protein is nammed the | + | The yellow chromoprotein used to create this fusion protein is nammed the fwYellow, a yellow chromoprotein ([http://parts.igem.org/Part:BBa_K1033910 BBa_K1033910] developed by iGEM13_Uppsala). |
| - | Homology modeling via Swiss-Model for the first chromoprotein gives us a nearly exact match with amFP486 which is a cyan fluorescent protein from Anemonia majano (the same organism as | + | Homology modeling via Swiss-Model for the first chromoprotein gives us a nearly exact match with amFP486 which is a cyan fluorescent protein from ''Anemonia majano'' (the same organism as the Uppsala's biobrick). Only 3 amino-acids change between the sequence of the two chromoproteins. These amino-acids are in the core of the protein. We can therefore hypothesize that at least three positions can be modified to change the emission spectrum of the chromoprotein. |
| - | [[File:Paris_Saclay_YellowChromoSwissMod.png|center| | + | [[File:Paris_Saclay_YellowChromoSwissMod.png|center|660px]] |
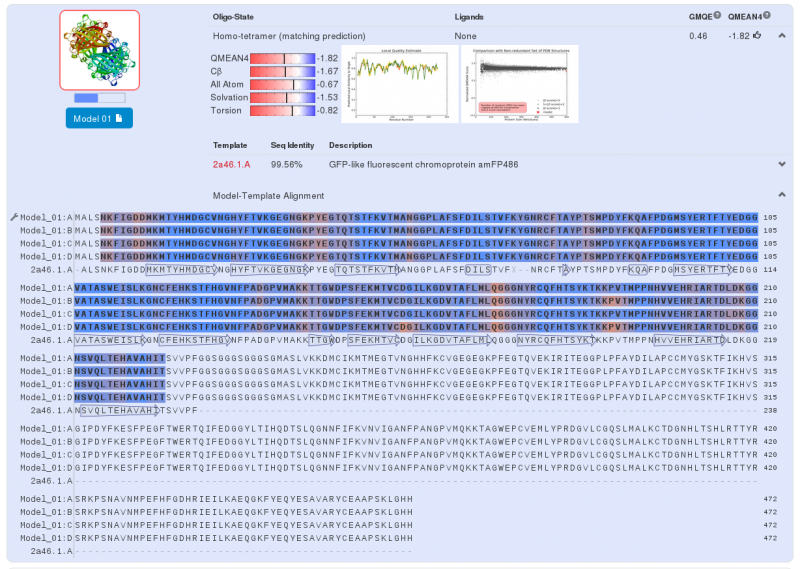
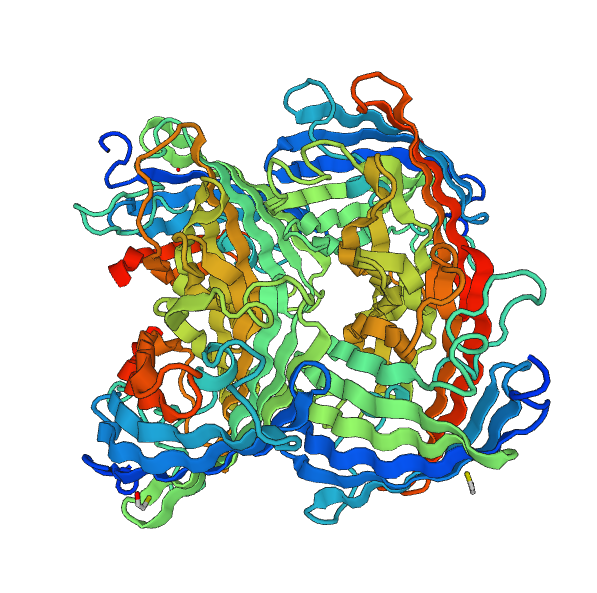
Note that this chromoprotein is a homotetramer. What might be able to raise problems is if homotetramerisation is essential to the functionality of the protein. The literature '''[3]''' tells us that it does not appear to be the case: fusion proteins have already been created with chromoproteins, which were homo-multimeric and the protein fusions were completely functional. | Note that this chromoprotein is a homotetramer. What might be able to raise problems is if homotetramerisation is essential to the functionality of the protein. The literature '''[3]''' tells us that it does not appear to be the case: fusion proteins have already been created with chromoproteins, which were homo-multimeric and the protein fusions were completely functional. | ||
| - | [[File:Paris_Saclay_YellowChromoSwissModStruct.png|center]] | + | [[File:Paris_Saclay_YellowChromoSwissModStruct.png|center|250px]] |
We predict that in our project, an homotetrameric state is not important to the functionality of our fusion protein. It is believed that the light emission will be less. | We predict that in our project, an homotetrameric state is not important to the functionality of our fusion protein. It is believed that the light emission will be less. | ||
| Line 33: | Line 25: | ||
We can do the same analysis for the blue chromoprotein. | We can do the same analysis for the blue chromoprotein. | ||
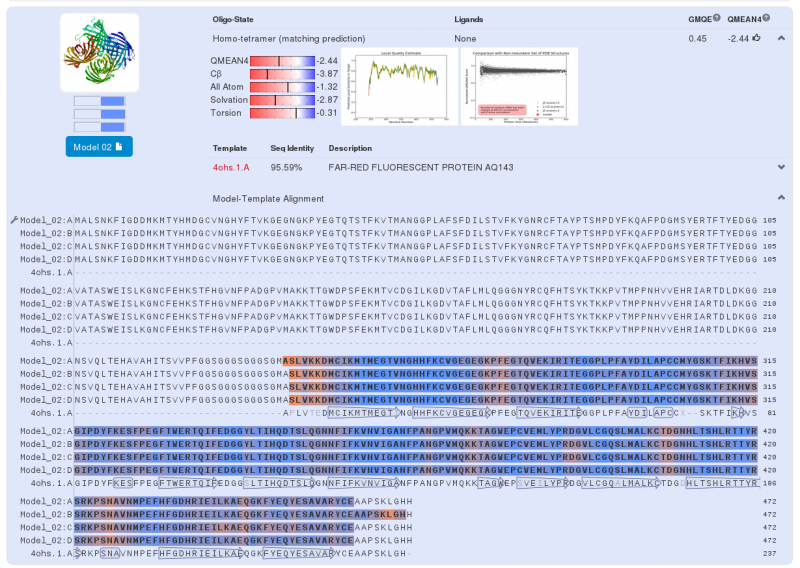
| - | For the second part of the fusion protein we use to the aeBlue which is a blue chromoprotein (Bba_K1033929 | + | For the second part of the fusion protein we use to the aeBlue which is a blue chromoprotein ([http://parts.igem.org/Part:BBa_K1033929 Bba_K1033929] developed by iGEM13_Uppsala). |
| - | Swiss-Model gives us a model for the second part | + | Swiss-Model gives us a model for the second part: a near perfect match between our protein sequence and chromoprotein (far-red fluorescent protein AQ143). There is a 95.59% similarity between the two sequences, the major differences are present at the C-terminus and N-terminus. As for the Yellow chromoprotein, three amino acids of the core protein are not the same. |
| - | [[File:Paris_Saclay_BlueChromoSwissMod.png|center| | + | [[File:Paris_Saclay_BlueChromoSwissMod.png|center|660px]] |
| - | Note that this chromoprotein is a homotetramer. | + | Note that this chromoprotein is also a homotetramer. |
| - | [[File:Paris_Saclay_BlueChromoSwissModStruct.png|center]] | + | [[File:Paris_Saclay_BlueChromoSwissModStruct.png|center|400px]] |
| - | Modeling with Swiss-Model does not provide information on the overall structure of our chromoprotein. Nonetheless, it gives us a very good approach to the structure of two chromoproteins which make up our fusion protein. | + | Modeling with Swiss-Model does not provide information on the overall structure of our chromoprotein. Nonetheless, it gives us a very good approach to the structure of the two chromoproteins which make up our fusion protein. |
==The last part – Global modelisation:== | ==The last part – Global modelisation:== | ||
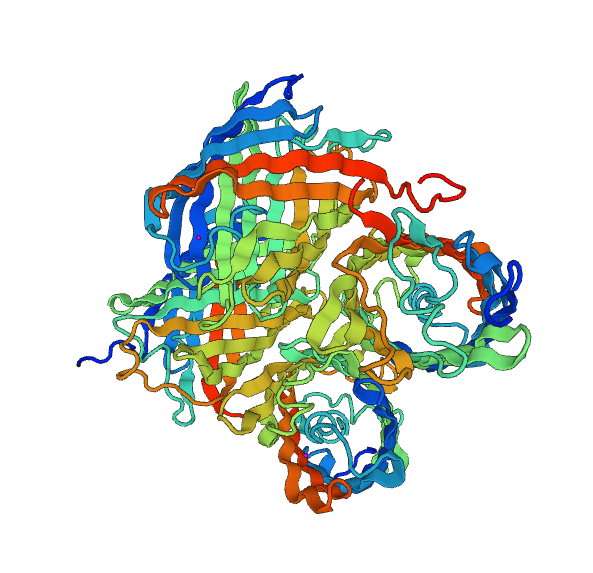
| - | We could make a much more complex modelisation for the overall structure of the fusion protein, we think it is unnecessary. Therefore, | + | We could make a much more complex modelisation for the overall structure of the fusion protein, we think it is unnecessary. Therefore, to have a global vision of our protein we will use the RaptorX model. |
| - | [[File:Paris_Saclay_Fusion-Chromoprotein-Struct.png|center| | + | [[File:Paris_Saclay_Fusion-Chromoprotein-Struct.png|center|400px]] |
Once this new model was obtained, we performed structural alignments and we discovered the structure of the two chromoproteins, so it seems that the linker between the two proteins is not sufficient to induce stress on the structures of our fusion protein. | Once this new model was obtained, we performed structural alignments and we discovered the structure of the two chromoproteins, so it seems that the linker between the two proteins is not sufficient to induce stress on the structures of our fusion protein. | ||
| Line 54: | Line 46: | ||
Obviously many biases have been introduced during this modeling and the conception of the fusion protein. This modelisation is closer to a structural analysis that a complex protein modeling. | Obviously many biases have been introduced during this modeling and the conception of the fusion protein. This modelisation is closer to a structural analysis that a complex protein modeling. | ||
The assumptions used in the design of this chromoprotein (fusion protein) does not seem to be so far from reality. We found similar cases in scientific publications, which are perfectly functional. | The assumptions used in the design of this chromoprotein (fusion protein) does not seem to be so far from reality. We found similar cases in scientific publications, which are perfectly functional. | ||
| - | We | + | We suggest several hypothesis to explain the failure of our experience. The main hypothesis is an error in the design of the nucleotide sequence that we requested to synthesize |
==References== | ==References== | ||
Latest revision as of 00:54, 16 October 2014

Contents |
Fusion Protein
Here we will discuss the structural modeling of the fusion protein that we had planned in order to obtain a green chromoprotein. We used the bioinformatics tools developed by the Swiss Institute of Bioinformatics (Swiss-Model [1]) and by the Xu group of Chicago University (RaptorX [2]).
Swiss-Model has given us three highly probable models. Two models for the first part of our Fusion Protein and another for second part of our protein. We chose the first model for the yellow chromoprotein (1st part) and the second model for the blue chromoprotein (2nde part).
The first part – Yellow chromoprotein:
The yellow chromoprotein used to create this fusion protein is nammed the fwYellow, a yellow chromoprotein ([http://parts.igem.org/Part:BBa_K1033910 BBa_K1033910] developed by iGEM13_Uppsala). Homology modeling via Swiss-Model for the first chromoprotein gives us a nearly exact match with amFP486 which is a cyan fluorescent protein from Anemonia majano (the same organism as the Uppsala's biobrick). Only 3 amino-acids change between the sequence of the two chromoproteins. These amino-acids are in the core of the protein. We can therefore hypothesize that at least three positions can be modified to change the emission spectrum of the chromoprotein.
Note that this chromoprotein is a homotetramer. What might be able to raise problems is if homotetramerisation is essential to the functionality of the protein. The literature [3] tells us that it does not appear to be the case: fusion proteins have already been created with chromoproteins, which were homo-multimeric and the protein fusions were completely functional.
We predict that in our project, an homotetrameric state is not important to the functionality of our fusion protein. It is believed that the light emission will be less.
The second part – blue chromoprotein:
We can do the same analysis for the blue chromoprotein. For the second part of the fusion protein we use to the aeBlue which is a blue chromoprotein ([http://parts.igem.org/Part:BBa_K1033929 Bba_K1033929] developed by iGEM13_Uppsala). Swiss-Model gives us a model for the second part: a near perfect match between our protein sequence and chromoprotein (far-red fluorescent protein AQ143). There is a 95.59% similarity between the two sequences, the major differences are present at the C-terminus and N-terminus. As for the Yellow chromoprotein, three amino acids of the core protein are not the same.
Note that this chromoprotein is also a homotetramer.
Modeling with Swiss-Model does not provide information on the overall structure of our chromoprotein. Nonetheless, it gives us a very good approach to the structure of the two chromoproteins which make up our fusion protein.
The last part – Global modelisation:
We could make a much more complex modelisation for the overall structure of the fusion protein, we think it is unnecessary. Therefore, to have a global vision of our protein we will use the RaptorX model.
Once this new model was obtained, we performed structural alignments and we discovered the structure of the two chromoproteins, so it seems that the linker between the two proteins is not sufficient to induce stress on the structures of our fusion protein.
Obviously many biases have been introduced during this modeling and the conception of the fusion protein. This modelisation is closer to a structural analysis that a complex protein modeling. The assumptions used in the design of this chromoprotein (fusion protein) does not seem to be so far from reality. We found similar cases in scientific publications, which are perfectly functional. We suggest several hypothesis to explain the failure of our experience. The main hypothesis is an error in the design of the nucleotide sequence that we requested to synthesize
References
[1] [http://swissmodel.expasy.org/ Swiss-Model server]
[2] Protein Structure and Function Prediction, [http://raptorx.uchicago.edu/ RaptorX server], Chicago university
[3] [http://nar.oxfordjournals.org/content/33/5/e49.full.pdf+html A dual-fluorescence reporter system for high-throughput clone characterization and selection by cell sorting] - Juno Choe, Haiwei H. Guo and Ger van den Engh, Nucleic Acids Research, 2005, Vol. 33, No. 5
 "
"