Team:Paris Saclay/Notebook/September/2
From 2014.igem.org
(→B-Contruction of the fusion protein) |
m (→Tuesday 2nd September) |
||
| (13 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{Team:Paris_Saclay/notebook_header}} | ||
| + | =Tuesday 2nd September= | ||
| + | |||
==Labwork== | ==Labwork== | ||
| - | === | + | ===Contruction of the fusion protein=== |
'' By Hv'' | '' By Hv'' | ||
| Line 20: | Line 23: | ||
We first did a liquid culture in 5 mL of LB. | We first did a liquid culture in 5 mL of LB. | ||
| - | === | + | ===Lemon scent=== |
''By Melanie'' | ''By Melanie'' | ||
| - | Limonene synthase | + | ====Limonene synthase==== |
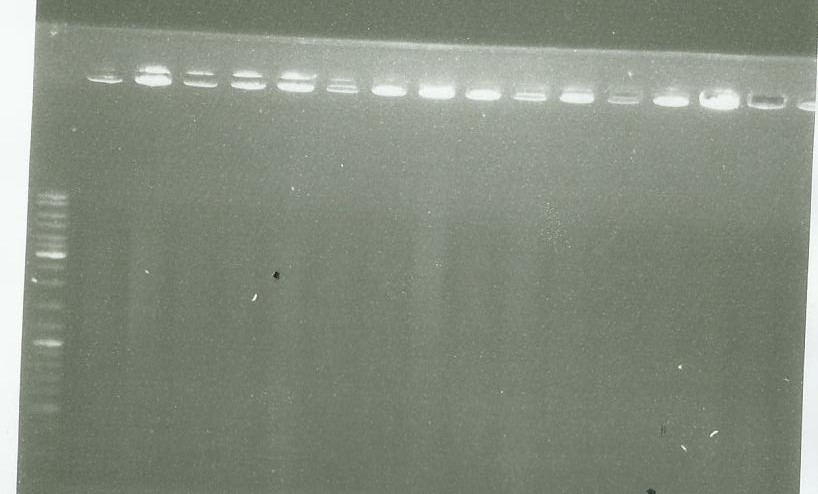
| - | Migration of the PCR done [https://2014.igem.org/Team:Paris_Saclay/Notebook/September/1 yesterday] | + | Migration of the PCR done [https://2014.igem.org/Team:Paris_Saclay/Notebook/September/1#Limone_synthase yesterday] |
| - | + | [[File:PCR pPS5 CAD + PCR LSpGMETe 0209.jpg|500px|center]] | |
| + | [[File:PCR clone LS pGMETe (12-25) 0209.jpg|500px|center]] | ||
| + | picture 1 | ||
well 1= ladder | well 1= ladder | ||
well 2-6 = pPS5 | well 2-6 = pPS5 | ||
other well = LS PCR | other well = LS PCR | ||
| + | |||
| + | picture 2 = LS PCR | ||
As we see, there are nothing on our gel so we conclude that we failed to clone LS in pGMETeasy | As we see, there are nothing on our gel so we conclude that we failed to clone LS in pGMETeasy | ||
| - | pPPS5 | + | ====pPPS5==== |
the photo in the same as previously: We saw a band but we will check another time by PCR to be sure. | the photo in the same as previously: We saw a band but we will check another time by PCR to be sure. | ||
| - | pPS3 and pPS4 | + | ====pPS3 and pPS4==== |
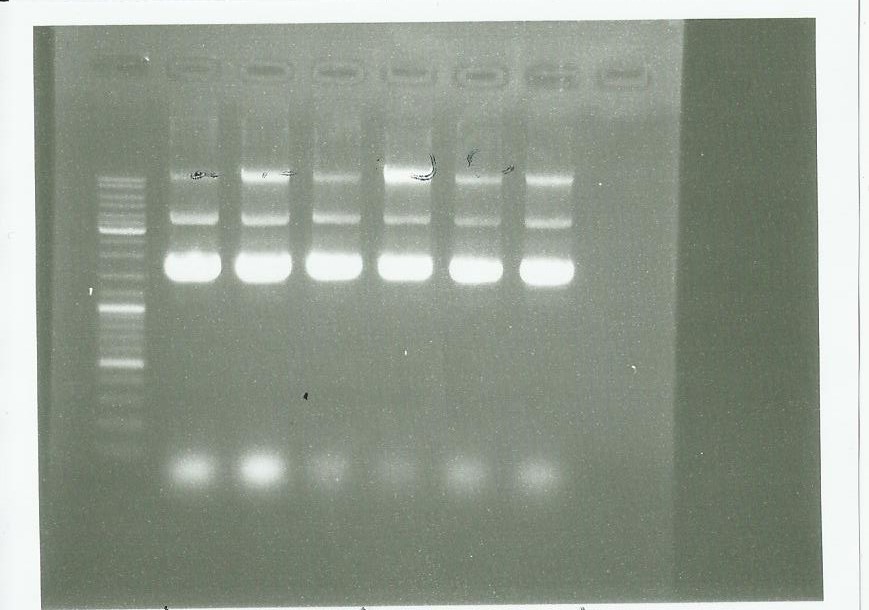
Plasmid extraction using the plasmid DNA extraction kit and electrophoresis: | Plasmid extraction using the plasmid DNA extraction kit and electrophoresis: | ||
| - | [ | + | [[File:Extraction, pPS3 pPS4 0209.jpg|500px|center]] |
We conclude that we have done a good manipulation. | We conclude that we have done a good manipulation. | ||
| + | ====Limonene synthase PCR==== | ||
| - | |||
As we saw that we don't have anything for LS, We try to do other PCR from the Biobrick (BBA762100). We use two enzyme: Dream taq and Vent taq | As we saw that we don't have anything for LS, We try to do other PCR from the Biobrick (BBA762100). We use two enzyme: Dream taq and Vent taq | ||
| + | |||
| + | {| class="wikitable centre" width="50%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | component | ||
| + | ! scope=col | volume | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |42μl | ||
| + | |- | ||
| + | |buffer | ||
| + | |5μl | ||
| + | |- | ||
| + | |dNTPs | ||
| + | |1μl | ||
| + | |- | ||
| + | |iPS66 | ||
| + | |1μl | ||
| + | |- | ||
| + | |iPS67 | ||
| + | |1μl | ||
| + | |- | ||
| + | |DNA | ||
| + | |0.5μl | ||
| + | |- | ||
| + | |enzyme | ||
| + | |0.5μl | ||
| + | |} | ||
| + | |||
| + | Tubes were placed in PCR machine with the following parameters. | ||
| + | |||
| + | {| class="wikitable centre" width="80%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | Cycle step | ||
| + | ! scope=col | Temperature | ||
| + | ! scope=col | Time | ||
| + | ! scope=col | Cycle | ||
| + | |- | ||
| + | | width="25%" | | ||
| + | Initial denaturation | ||
| + | | width="25%" | | ||
| + | 94°C | ||
| + | | width="25%" | | ||
| + | 1 min | ||
| + | | width="25%" | | ||
| + | 1 | ||
| + | |- | ||
| + | | Denaturation | ||
| + | | 94°C | ||
| + | | 30 s | ||
| + | | 25 | ||
| + | |- | ||
| + | | Annealing | ||
| + | | 50°C | ||
| + | | 25 s | ||
| + | | 25 | ||
| + | |- | ||
| + | | Extension | ||
| + | | 72°C | ||
| + | | 1 min | ||
| + | | 25 | ||
| + | |- | ||
| + | | Final extension | ||
| + | | 72°C | ||
| + | | 10 min | ||
| + | | 1 | ||
| + | |- | ||
| + | | Final extension | ||
| + | | 8°C | ||
| + | | hold | ||
| + | | 1 | ||
| + | |} | ||
| + | |||
| + | ====pPS5==== | ||
| + | To have more plasmid We do some bacteria culture (pPS5) | ||
| + | |||
| + | ==Photo of the Day== | ||
| + | [[File:Paris Saclay 16_september.jpg|600px|center]] | ||
| + | |||
| + | {{Team:Paris_Saclay/notebook_footer}} | ||
Latest revision as of 12:36, 14 October 2014
Contents |
Tuesday 2nd September
Labwork
Contruction of the fusion protein
By Hv
We can see that the blue colonies of E.coli are not blue in a simple petri dish with ampicillin. It's a fail. So, to figure our problem we decided to do a sequencing of our plasmids.
We chose 6 colonies from an Xgal-IPTG petri dish:
- 1 completely blue
- 2 blue with a white ring around the blue
- 3 white colony
PS: we found that there is 2 types of blue colonies: completely blue and blue with a white ring around the blue)
We first did a liquid culture in 5 mL of LB.
Lemon scent
By Melanie
Limonene synthase
Migration of the PCR done yesterday
picture 1 well 1= ladder well 2-6 = pPS5 other well = LS PCR
picture 2 = LS PCR As we see, there are nothing on our gel so we conclude that we failed to clone LS in pGMETeasy
pPPS5
the photo in the same as previously: We saw a band but we will check another time by PCR to be sure.
pPS3 and pPS4
Plasmid extraction using the plasmid DNA extraction kit and electrophoresis:
We conclude that we have done a good manipulation.
Limonene synthase PCR
As we saw that we don't have anything for LS, We try to do other PCR from the Biobrick (BBA762100). We use two enzyme: Dream taq and Vent taq
| component | volume |
|---|---|
| H2O | 42μl |
| buffer | 5μl |
| dNTPs | 1μl |
| iPS66 | 1μl |
| iPS67 | 1μl |
| DNA | 0.5μl |
| enzyme | 0.5μl |
Tubes were placed in PCR machine with the following parameters.
| Cycle step | Temperature | Time | Cycle |
|---|---|---|---|
|
Initial denaturation |
94°C |
1 min |
1 |
| Denaturation | 94°C | 30 s | 25 |
| Annealing | 50°C | 25 s | 25 |
| Extension | 72°C | 1 min | 25 |
| Final extension | 72°C | 10 min | 1 |
| Final extension | 8°C | hold | 1 |
pPS5
To have more plasmid We do some bacteria culture (pPS5)
 "
"