Team:Colombia/Classes
From 2014.igem.org
Camilog137 (Talk | contribs) |
Camilog137 (Talk | contribs) |
||
| (61 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{Http://2014.igem.org/Team:Colombia | + | {{Http://2014.igem.org/Team:Colombia}} |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<html> | <html> | ||
| + | <div class="span11" style="text-align: justify;"> | ||
| - | < | + | <center><b> <font size="24"> |
| - | + | <br><br> | |
| - | + | Classes | |
| - | + | </font><br> | |
| - | + | <h1 class="curs1"><font size="6"> | |
| - | + | Sharing our knowledge | |
| - | + | </font></h1></b></center> | |
| - | + | ||
| - | + | ||
| - | + | <p align="justify"> | |
| - | + | <font size=3>Our team is made up of students from different areas of knowlegde, from design to engineering. We consider the only thing you need in order to be part of us is motivation. This is why we made a series of classes given by professors and advanced senior students where everyone can learn about everything that the project is going to be about. So our classes started as basic as cellular biology, passing through matlab, differential equations, genetic circuits and lab basics, such as restriction enzymes and PCR. Here you can check our presentations, and there is to stand out that all this training was made by students from iGEM 2012 and 2013, so they can pass all they experience to the new members of the group. | |
| - | + | <br><br> | |
| - | + | We made PopQuizzeess and All! :)</font> | |
| - | + | </p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <br> | |
| - | + | <font color="#8A0808" size="5" >Basic Biology</font> | |
| - | + | <br> | |
| - | + | ||
| - | + | ||
</html> | </html> | ||
| + | [[File:Col_Biology.pdf|left]] | ||
| + | [[File:Col_Biology.jpg|left|300 px]] | ||
| + | <br> | ||
| + | <br> | ||
| + | <html><p> | ||
| + | <font size="3" >We have students from engineering backgrounds who have not study biology. With this class we started by the basic differences between procaryotes and eucaryotes and learnt all the cell biology required to understand systems biology processes, . </font> | ||
| + | </p> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br><br><br> | ||
| + | <font color="#8A0808" size="5" >Molecular Biology</font> | ||
| + | <br> | ||
| + | </html> | ||
| + | [[File:Col_MolBol.pdf|left]] | ||
| + | [[File:Col_MOlBiol.jpg|left|300 px]] | ||
| + | <br> | ||
| + | <br> | ||
| + | <html><p> | ||
| + | <font size="3" >Once the biology basics were stablished we continue to learn molecular biology. We learn about DNA structure and manipulation. We understood deeply PCR and Restriction enzymes</font> | ||
| + | </p> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
<br><br> | <br><br> | ||
| + | <font color="#8A0808" size="5" >Quorum Sensing</font> | ||
| + | <br> | ||
| + | </html> | ||
| + | [[File:Quorum Sensing.pdf|left]] | ||
| + | [[File:Quorum Sensing.jpg|left|300 px]] | ||
| + | <br> | ||
| + | <br> | ||
| + | <html><p> | ||
| + | <font size="3" >With this class we learnt the basic topics in bacterial quorum sensing and how they talk to each other. For instance the concepts about AHL, Lux-like genes and the effect of population density were examined. In addition we saw some applications of bacterial quorum sensing in genetic constructs, illustrating this with well known examples of bacterial signaling in synthetic biology.</font> | ||
</p> | </p> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <font color="#8A0808" size="5" >Differential equations</font> | ||
| + | <br> | ||
| + | </html> | ||
| + | [[File:Differential Equations.pdf|left]] | ||
| + | [[File:Differential Equations.jpg|left|300 px]] | ||
| + | <br> | ||
| + | <br> | ||
| + | <html><p> | ||
| + | <font size="3" >For those of us who did not have a mathematical background we made this class to understand what a differential equation is. We learnt how to made equations from biological processes and that there were two ways to find their solutions: Analytically and Numerically</font> | ||
| + | </p> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <font color="#8A0808" size="5" >Mathematical models</font> | ||
| + | <br> | ||
| + | </html> | ||
| + | [[File:Mathematical Models.pdf|left]] | ||
| + | [[File:MathModels.jpg|left|300 px]] | ||
| + | <br> | ||
| + | <br> | ||
| + | <html><p> | ||
| + | <font size="3" >One of the main sections of iGEM are mathematical models, with this class we learnt how they are made. Concepts like the law of mass action and the mass conservation principle were discussed in order to understand the Michaelis Menten and the Hill's expression demonstrations. We studied a way to find the parameters for the model developed by the </html> [https://2012.igem.org/Team:Colombia/Modeling/Paramterers Team in 2012]<html> </font> | ||
| + | </p> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <center> | ||
| + | <div class="button-fill orange" ><div class="button-text">Back Notebook</div><div class="button-inside"><div class="inside-text"><a style="text-decoration: none; background-color: none; color: red;" href="https://2014.igem.org/Team:Colombia/Notebook">Go! </a></div></div></div> | ||
| + | </center> | ||
| - | + | <br><br><br><br><br><br> | |
| - | + | </div> | |
| - | + | ||
| - | + | <script> | |
| - | - | + | $(".button-fill").hover(function () { |
| - | + | $(this).children(".button-inside").addClass('full'); | |
| - | + | }, function() { | |
| - | + | $(this).children(".button-inside").removeClass('full'); | |
| - | + | }); | |
| - | + | //@ sourceURL=pen.js | |
| - | + | </script> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | </ | + | |
Latest revision as of 01:47, 14 October 2014
Classes
Sharing our knowledge
Our team is made up of students from different areas of knowlegde, from design to engineering. We consider the only thing you need in order to be part of us is motivation. This is why we made a series of classes given by professors and advanced senior students where everyone can learn about everything that the project is going to be about. So our classes started as basic as cellular biology, passing through matlab, differential equations, genetic circuits and lab basics, such as restriction enzymes and PCR. Here you can check our presentations, and there is to stand out that all this training was made by students from iGEM 2012 and 2013, so they can pass all they experience to the new members of the group.
We made PopQuizzeess and All! :)
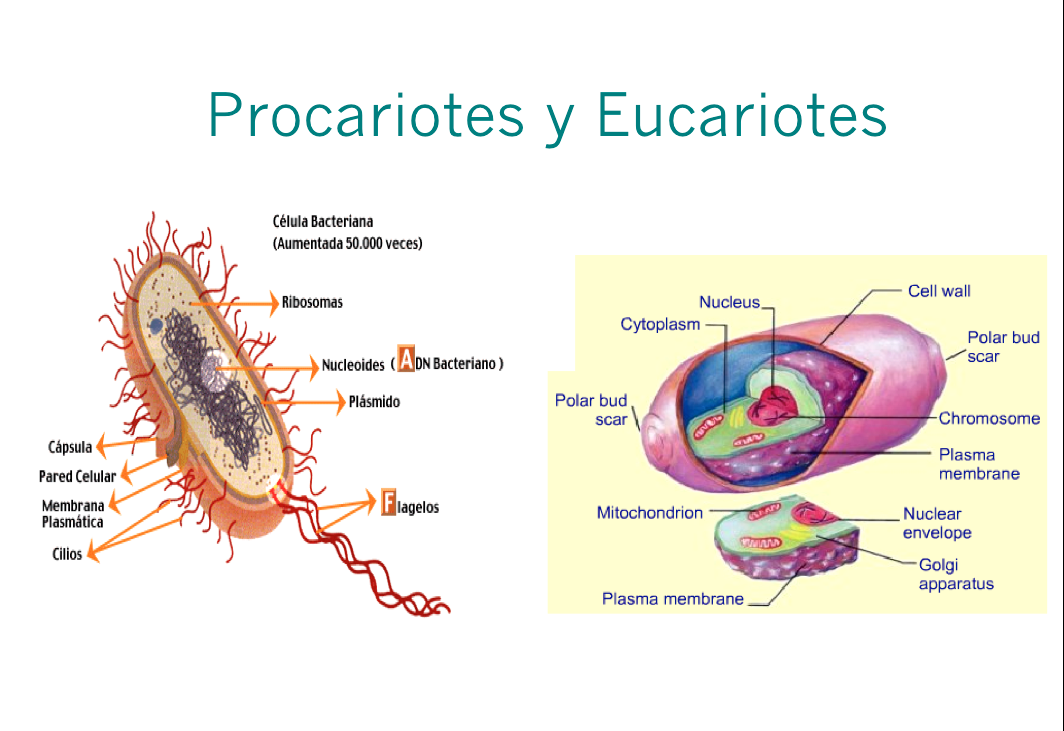
Basic Biology
File:Col Biology.pdf
We have students from engineering backgrounds who have not study biology. With this class we started by the basic differences between procaryotes and eucaryotes and learnt all the cell biology required to understand systems biology processes, .
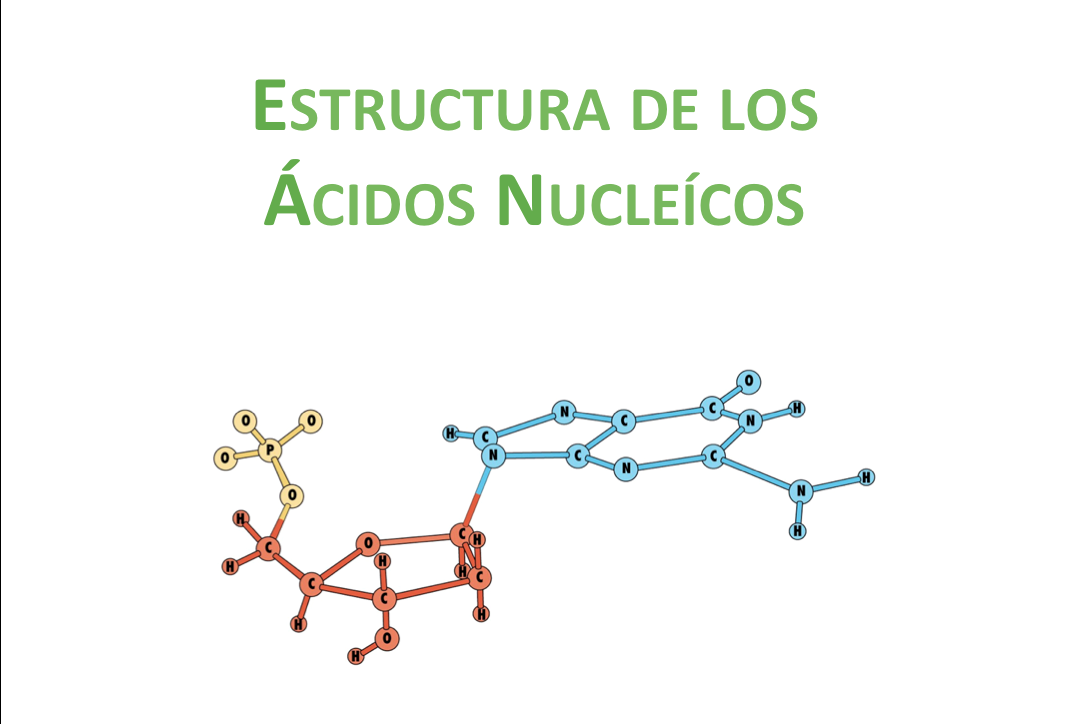
Molecular Biology
File:Col MolBol.pdf
Once the biology basics were stablished we continue to learn molecular biology. We learn about DNA structure and manipulation. We understood deeply PCR and Restriction enzymes
Quorum Sensing
File:Quorum Sensing.pdf
With this class we learnt the basic topics in bacterial quorum sensing and how they talk to each other. For instance the concepts about AHL, Lux-like genes and the effect of population density were examined. In addition we saw some applications of bacterial quorum sensing in genetic constructs, illustrating this with well known examples of bacterial signaling in synthetic biology.
Differential equations
File:Differential Equations.pdf
For those of us who did not have a mathematical background we made this class to understand what a differential equation is. We learnt how to made equations from biological processes and that there were two ways to find their solutions: Analytically and Numerically
Mathematical models
File:Mathematical Models.pdf
One of the main sections of iGEM are mathematical models, with this class we learnt how they are made. Concepts like the law of mass action and the mass conservation principle were discussed in order to understand the Michaelis Menten and the Hill's expression demonstrations. We studied a way to find the parameters for the model developed by the Team in 2012
 "
"