Team:Paris Saclay/Project/Odor-free ecoli
From 2014.igem.org
(Created page with "{{Team:Paris_Saclay/default_header}} =Odor-free E.coli chassis= ==Sub title== Lorem ipsum dolor sit amet, consectetur adipiscing elit. In vestibulum ac lacus luctus semper. Etiam...") |
(→Countdown) |
||
| (26 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{Team:Paris_Saclay/ | + | {{Team:Paris_Saclay/project_header}} |
| - | = | + | =Remove the bad smell of ''E.coli''= |
| - | == | + | ==Introduction== |
| - | + | ||
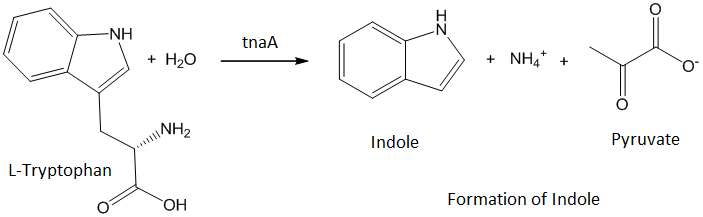
| - | + | ''Escherichia coli'' stinks because of the ''tnaA'' gene which produces an enzyme that transforms the L-tryptophan into indole, responsible for the stench. If we want our lemon to smell like one, we have to delete this gene. | |
| + | [[File:Paris_Saclay_IndoleFormation.png|500px|center]] | ||
| + | |||
| + | ==The Principle of the construction== | ||
| + | |||
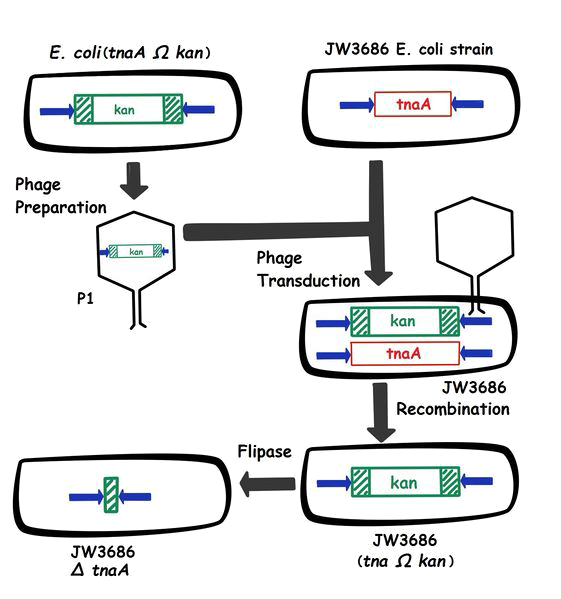
| + | In the lab, we already had a strain in which the tnaA was replaced by a kanamycin resistance, but this strain was too modified to be used for our project. So we switched the tnaA sequence with the kanamycin resistance in our bacterium by phage transduction. Transduction is the transfer of bacterial DNA from one bacterium (the donor) to another (the recipient) by a bacteriophage P1. | ||
| + | |||
| + | At first, the modified bacteria (cell donor) are infected by P1 phage. Transducing bacteriophage particles are formed in the donor bacterial cells during the phage growth. The bacterial chromosome is broken up into small fragments (approximately the same length as the P1 DNA). The phage particles mistakenly incorporate the kanamycin resistance from the donor in place of phage DNA. | ||
| + | |||
| + | Once packaged, the transducing bacterial DNA is delivered to recipient bacterium (containing the tnaA sequence) as if it were phage DNA (linear DNA molecules are injected through the wall and cytoplasmic membranes into the cell). Then, there is homologous recombination with the DNA of the recipient bacterium; the bacterium acquires the kanamycin resistance gene of the donor. The transduction is complete, the tnaA sequence has been correctly replaced by the kanamycin resistance. Then, we verified by screening on petri dishes with kanamycin. | ||
| + | |||
| + | After the recombination, we used a flippase. The flippase recognizes small sequences called Flippase recognition target (frt), and catalyzes a site-specific recombination reaction between the two frt sites. In this case, it is used to remove the kanamycin resistance gene. | ||
| + | |||
| + | The remaining bacterium doesn't smell at all. | ||
| + | |||
| + | [[File:Paris Saclay Odor Free coli.png|500px|center]] | ||
| - | |||
{{Team:Paris_Saclay/default_footer}} | {{Team:Paris_Saclay/default_footer}} | ||
Latest revision as of 06:24, 17 October 2014

Remove the bad smell of E.coli
Introduction
Escherichia coli stinks because of the tnaA gene which produces an enzyme that transforms the L-tryptophan into indole, responsible for the stench. If we want our lemon to smell like one, we have to delete this gene.
The Principle of the construction
In the lab, we already had a strain in which the tnaA was replaced by a kanamycin resistance, but this strain was too modified to be used for our project. So we switched the tnaA sequence with the kanamycin resistance in our bacterium by phage transduction. Transduction is the transfer of bacterial DNA from one bacterium (the donor) to another (the recipient) by a bacteriophage P1.
At first, the modified bacteria (cell donor) are infected by P1 phage. Transducing bacteriophage particles are formed in the donor bacterial cells during the phage growth. The bacterial chromosome is broken up into small fragments (approximately the same length as the P1 DNA). The phage particles mistakenly incorporate the kanamycin resistance from the donor in place of phage DNA.
Once packaged, the transducing bacterial DNA is delivered to recipient bacterium (containing the tnaA sequence) as if it were phage DNA (linear DNA molecules are injected through the wall and cytoplasmic membranes into the cell). Then, there is homologous recombination with the DNA of the recipient bacterium; the bacterium acquires the kanamycin resistance gene of the donor. The transduction is complete, the tnaA sequence has been correctly replaced by the kanamycin resistance. Then, we verified by screening on petri dishes with kanamycin.
After the recombination, we used a flippase. The flippase recognizes small sequences called Flippase recognition target (frt), and catalyzes a site-specific recombination reaction between the two frt sites. In this case, it is used to remove the kanamycin resistance gene.
The remaining bacterium doesn't smell at all.
 "
"