Team:Paris Saclay/Notebook/August/20
From 2014.igem.org
(→D- Lemon scent) |
m (→Photo of the Day) |
||
| (31 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Paris_Saclay/notebook_header}} | {{Team:Paris_Saclay/notebook_header}} | ||
=Wednesday 20th August= | =Wednesday 20th August= | ||
| - | { | + | ==Lab work== |
| + | ====Petri dishes==== | ||
| + | ''by Laetitia'' | ||
| + | |||
| + | {| class="wikitable centre" width="25%" | ||
| + | |+ 10 dishes with: | ||
| + | |- | ||
| + | ! scope=col | Antibiotic | ||
| + | ! scope=col | Concentration | ||
| + | |- | ||
| + | |XGal | ||
| + | |80μg/ml | ||
| + | |- | ||
| + | |IPTG | ||
| + | |2*10<sup>-3</sup> | ||
| + | |- | ||
| + | |Ampicillin | ||
| + | |10<sup>-3</sup> | ||
| + | |} | ||
| + | |||
| + | ===Construction of the fusion protein (color)=== | ||
| + | ====PCR==== | ||
| + | |||
| + | {| class="wikitable centre" width="50%" | ||
| + | |+ PCR of chromoprotein | ||
| + | |- | ||
| + | ! scope=col | components | ||
| + | ! scope=col | volumes | ||
| + | |- | ||
| + | |DNA | ||
| + | |.5µl | ||
| + | |- | ||
| + | |GoTaq buffer 5X | ||
| + | |10μl | ||
| + | |- | ||
| + | |DMSO | ||
| + | |1.5µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |22.75µl | ||
| + | |- | ||
| + | |MgCl<sub>2</sub> | ||
| + | |4µl | ||
| + | |- | ||
| + | |dNTP | ||
| + | |1µl | ||
| + | |- | ||
| + | |iPS83 | ||
| + | |5µl | ||
| + | |- | ||
| + | |iPS84 | ||
| + | |5µl | ||
| + | |- | ||
| + | |GoTaq polymerase | ||
| + | |.25µl | ||
| + | |} | ||
| + | |||
| + | {| class="wikitable centre" width="50%" | ||
| + | |+ PCR cycle | ||
| + | |- | ||
| + | ! scope=col | step | ||
| + | ! scope=col | temperature (°C) | ||
| + | ! scope=col | time (min) | ||
| + | |- | ||
| + | |1 | ||
| + | |95 | ||
| + | |2 | ||
| + | |- | ||
| + | |2 | ||
| + | |95 | ||
| + | |.5 | ||
| + | |- | ||
| + | |3 | ||
| + | |58 | ||
| + | |.5 | ||
| + | |- | ||
| + | |4 | ||
| + | |72 | ||
| + | |1.75 | ||
| + | |- | ||
| + | |5 | ||
| + | |72 | ||
| + | |5 | ||
| + | |} | ||
| + | |||
| + | ====Cloning in pGEMTesay==== | ||
| + | ''by Sean'' | ||
| - | + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/ protocol] | |
| - | ===Bacterial culture=== | + | ===Lemon scent=== |
| + | |||
| + | ====Bacterial culture==== | ||
''by Laetitia'' | ''by Laetitia'' | ||
| + | [[File:20 08 bacterial culture.jpg| 500px|right]] | ||
Here are the results of the bacterial culture of yesterday | Here are the results of the bacterial culture of yesterday | ||
| - | |||
Bacteria with the pGEMTeasy without the DNA insert are blue and the bacteria with the pGEMTeasy and the DNA insert (PS, GS or LS gene) are white | Bacteria with the pGEMTeasy without the DNA insert are blue and the bacteria with the pGEMTeasy and the DNA insert (PS, GS or LS gene) are white | ||
| Line 27: | Line 115: | ||
Preculture 20ml of DH5a pPSI (apra 1/2000) | Preculture 20ml of DH5a pPSI (apra 1/2000) | ||
| + | ====CAD PCR==== | ||
| + | Today we received our synthetic gene CAD (cinnamyl alcohol deshydrogenase) :) | ||
| - | + | so we do a PCR (with GoTaq): | |
| - | + | ||
| - | + | {| class="wikitable centre" width="50%" | |
| + | |+ PCR of CAD | ||
| + | |- | ||
| + | ! scope=col | components | ||
| + | ! scope=col | volumes | ||
| + | |- | ||
| + | |DNA of CAD | ||
| + | |"very small amount" | ||
| + | |- | ||
| + | |GoTaq buffer 5X | ||
| + | |10μl | ||
| + | |- | ||
| + | |DMSO | ||
| + | |1.5µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |23.25µl | ||
| + | |- | ||
| + | |MgCl<sub>2</sub> | ||
| + | |4µl | ||
| + | |- | ||
| + | |dNTP | ||
| + | |1µl | ||
| + | |- | ||
| + | |iPS79 | ||
| + | |5µl | ||
| + | |- | ||
| + | |iPS80 | ||
| + | |5µl | ||
| + | |- | ||
| + | |GoTaq polymerase | ||
| + | |.25µl | ||
| + | |} | ||
| - | + | {| class="wikitable centre" width="50%" | |
| + | |+ PCR cycle | ||
| + | |- | ||
| + | ! scope=col | step | ||
| + | ! scope=col | temperature (°C) | ||
| + | ! scope=col | time (min) | ||
| + | |- | ||
| + | |1 | ||
| + | |95 | ||
| + | |2 | ||
| + | |- | ||
| + | |2 | ||
| + | |95 | ||
| + | |.5 | ||
| + | |- | ||
| + | |3 | ||
| + | |58 | ||
| + | |.5 | ||
| + | |- | ||
| + | |4 | ||
| + | |72 | ||
| + | |1.75 | ||
| + | |- | ||
| + | |5 | ||
| + | |72 | ||
| + | |5 | ||
| + | |} | ||
| - | + | ====CAD cloning in pGEMTeasy==== | |
| + | ''by Sean'' | ||
| - | + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/ protocol] | |
| - | + | ==== Electrophoresis of pPSI digested by PacI ==== | |
| + | ''by Eugene'' | ||
| - | + | We made an electrophoresis of pPSI to verify if the digestion made [https://2014.igem.org/Team:Paris_Saclay/Notebook/August/19#Digestion_of_PCR_results 19th August] has worked. It is useful before starting the purification of the plasmid. | |
| - | + | #non-digested pPSI 3µl | |
| + | #digested pPSI 3µl | ||
| + | #ladder 10µl | ||
| - | + | [[File:Paris_Saclay_140820.jpg|center]] | |
| - | + | ====Migration of pPS1 digested by PacI for the purification==== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ===Migration of pPS1 digested by PacI=== | + | |
''by Huang vu and Laetitia'' | ''by Huang vu and Laetitia'' | ||
| Line 72: | Line 211: | ||
The totality of the sample is filled (around 38µL) and we filled 20µl of ladder. Migration 100V | The totality of the sample is filled (around 38µL) and we filled 20µl of ladder. Migration 100V | ||
| - | After the migration, we want to purify the band corresponding of pPS1 digested by Pac1. While exposing the gel to UV, we will cut the band and save it inside an Eppendorf, | + | After the migration, we want to purify the band corresponding of pPS1 digested by Pac1. While exposing the gel to UV, we will cut the band and save it inside an Eppendorf (weight = 1.080g) , before the purification |
| - | ==== | + | ====Digestion of CAD and PMCS5 at 37°C==== |
| - | + | ''by Eugene'' | |
| - | + | {| class="wikitable centre" width="50%" | |
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | components | ||
| + | ! scope=col | volumes | ||
| + | |- | ||
| + | |CAD | ||
| + | |5 μl | ||
| + | |- | ||
| + | |10x buffer fastDigest | ||
| + | |1 μl | ||
| + | |- | ||
| + | |PacI | ||
| + | |0.5 µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |3.5 µl | ||
| + | |} | ||
| - | |||
| - | + | {| class="wikitable centre" width="50%" | |
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | components | ||
| + | ! scope=col | volumes | ||
| + | |- | ||
| + | |PMCS5 | ||
| + | |15 μl | ||
| + | |- | ||
| + | |10x buffer fastDigest | ||
| + | |2 μl | ||
| + | |- | ||
| + | |PacI | ||
| + | |0.5 µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |2.5 µl | ||
| + | |} | ||
| - | + | ==Photo of the Day== | |
| + | [[File:Paris Saclay 20_august.jpg|300px|center]] | ||
| - | + | '''Members present:''' | |
| + | * Instructors and advisors: Alice, Solenne and Sylvie. | ||
| + | * Students: Eugène, Hoang Vu, Laëtitia, Mélanie, Romain, Sean and Terry. | ||
| - | + | {{Team:Paris_Saclay/notebook_footer}} | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 15:08, 14 October 2014
Contents |
Wednesday 20th August
Lab work
Petri dishes
by Laetitia
| Antibiotic | Concentration |
|---|---|
| XGal | 80μg/ml |
| IPTG | 2*10-3 |
| Ampicillin | 10-3 |
Construction of the fusion protein (color)
PCR
| components | volumes |
|---|---|
| DNA | .5µl |
| GoTaq buffer 5X | 10μl |
| DMSO | 1.5µl |
| H2O | 22.75µl |
| MgCl2 | 4µl |
| dNTP | 1µl |
| iPS83 | 5µl |
| iPS84 | 5µl |
| GoTaq polymerase | .25µl |
| step | temperature (°C) | time (min) |
|---|---|---|
| 1 | 95 | 2 |
| 2 | 95 | .5 |
| 3 | 58 | .5 |
| 4 | 72 | 1.75 |
| 5 | 72 | 5 |
Cloning in pGEMTesay
by Sean
Lemon scent
Bacterial culture
by Laetitia
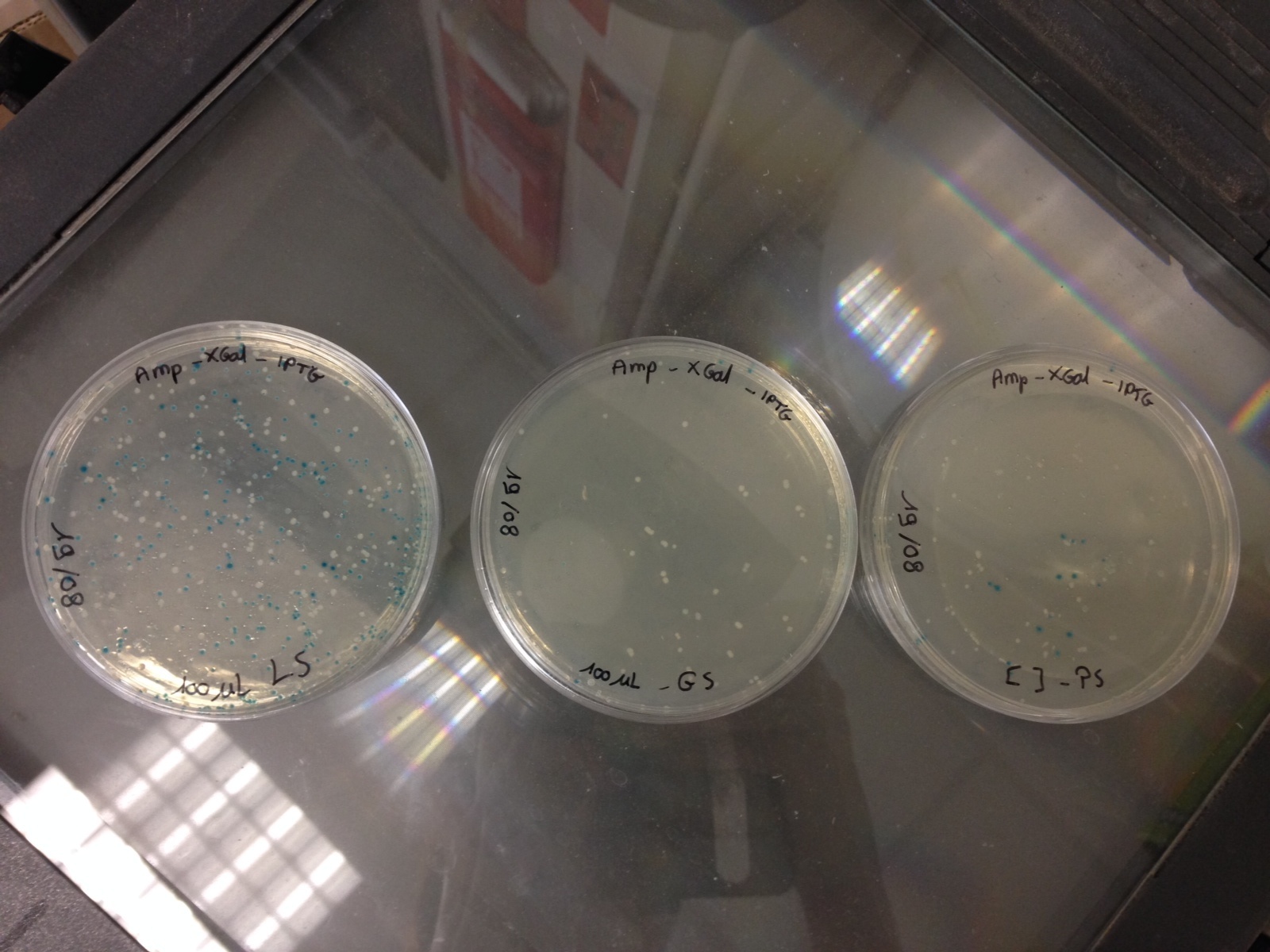
Here are the results of the bacterial culture of yesterday
Bacteria with the pGEMTeasy without the DNA insert are blue and the bacteria with the pGEMTeasy and the DNA insert (PS, GS or LS gene) are white
Then, we have chosen 3 white colonies for each DNA insert and we have launched 3 corresponding liquid cultures :
- 5mL LB + ampiciline (1/1000e)
- 1 white colony
The 9 cultures are incubated at 37°C
by Melanie
Preculture 20ml of DH5a pPSI (apra 1/2000)
CAD PCR
Today we received our synthetic gene CAD (cinnamyl alcohol deshydrogenase) :)
so we do a PCR (with GoTaq):
| components | volumes |
|---|---|
| DNA of CAD | "very small amount" |
| GoTaq buffer 5X | 10μl |
| DMSO | 1.5µl |
| H2O | 23.25µl |
| MgCl2 | 4µl |
| dNTP | 1µl |
| iPS79 | 5µl |
| iPS80 | 5µl |
| GoTaq polymerase | .25µl |
| step | temperature (°C) | time (min) |
|---|---|---|
| 1 | 95 | 2 |
| 2 | 95 | .5 |
| 3 | 58 | .5 |
| 4 | 72 | 1.75 |
| 5 | 72 | 5 |
CAD cloning in pGEMTeasy
by Sean
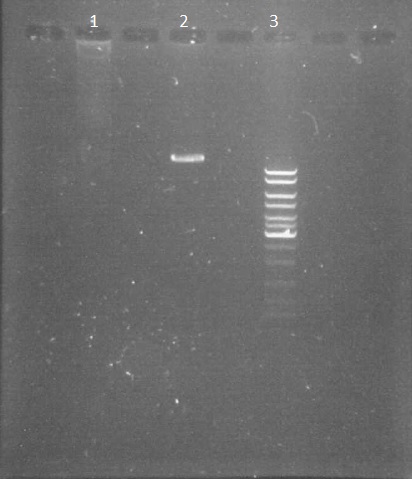
Electrophoresis of pPSI digested by PacI
by Eugene
We made an electrophoresis of pPSI to verify if the digestion made 19th August has worked. It is useful before starting the purification of the plasmid.
- non-digested pPSI 3µl
- digested pPSI 3µl
- ladder 10µl
Migration of pPS1 digested by PacI for the purification
by Huang vu and Laetitia
pPS1 previously digested by PacI is filled inside an agarose gel :
-1g agarose
-1mL TAE
-2 µL BET
The totality of the sample is filled (around 38µL) and we filled 20µl of ladder. Migration 100V
After the migration, we want to purify the band corresponding of pPS1 digested by Pac1. While exposing the gel to UV, we will cut the band and save it inside an Eppendorf (weight = 1.080g) , before the purification
Digestion of CAD and PMCS5 at 37°C
by Eugene
| components | volumes |
|---|---|
| CAD | 5 μl |
| 10x buffer fastDigest | 1 μl |
| PacI | 0.5 µl |
| H2O | 3.5 µl |
| components | volumes |
|---|---|
| PMCS5 | 15 μl |
| 10x buffer fastDigest | 2 μl |
| PacI | 0.5 µl |
| H2O | 2.5 µl |
Photo of the Day
Members present:
- Instructors and advisors: Alice, Solenne and Sylvie.
- Students: Eugène, Hoang Vu, Laëtitia, Mélanie, Romain, Sean and Terry.
 "
"