Team:Paris Saclay/Notebook/August/19
From 2014.igem.org
(→Lab work) |
m (→Bacterial culture) |
||
| (13 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
==Lab work== | ==Lab work== | ||
| - | === | + | ===Chromoprotein + Lemon scent=== |
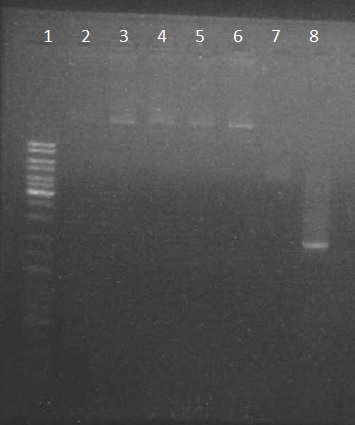
| + | Electrophoresis with the PpsI plasmide (obtain on thursday) + the chromoprotein PCR: | ||
| + | we obtain results as expected | ||
| + | |||
| + | #ladder 10µl | ||
| + | #pPSI 1.2µl | ||
| + | #pPSI 1.2µl | ||
| + | #pPSI 1.2µl | ||
| + | #pPSI 1.2µl | ||
| + | #pPSI 1.2µl | ||
| + | #p cola 1.2µl | ||
| + | #PCR chromoprotein 2µl | ||
| + | |||
| + | [[File:Paris_Saclay_140819.jpg]] | ||
| + | |||
| + | ===Lemon Scent=== | ||
==== Digestion of PCR results ==== | ==== Digestion of PCR results ==== | ||
| Line 64: | Line 79: | ||
|2 µl | |2 µl | ||
|} | |} | ||
| - | |||
{| class="wikitable centre" width="50%" | {| class="wikitable centre" width="50%" | ||
| Line 72: | Line 86: | ||
! scope=col | volumes | ! scope=col | volumes | ||
|- | |- | ||
| - | | | + | |Purified pPS1 |
|30 μl | |30 μl | ||
|- | |- | ||
| Line 84: | Line 98: | ||
|4 µl | |4 µl | ||
|} | |} | ||
| - | |||
====Purification of PCR products of limonen synthase==== | ====Purification of PCR products of limonen synthase==== | ||
| Line 100: | Line 113: | ||
We transformed 100µL of competent bacteria and we added the totality of the ligation product (pGEMTeasy containing LS or PS or GS) | We transformed 100µL of competent bacteria and we added the totality of the ligation product (pGEMTeasy containing LS or PS or GS) | ||
| - | ====Preparation of petri | + | ====Preparation of petri dishes==== |
''by Terry'' | ''by Terry'' | ||
| - | + | {| class="wikitable centre" width="25%" | |
| + | |+ 10 dishes with: | ||
| + | |- | ||
| + | ! scope=col | Antibiotic | ||
| + | ! scope=col | Concentration | ||
| + | |- | ||
| + | |XGal | ||
| + | |80μg/ml | ||
| + | |- | ||
| + | |IPTG | ||
| + | |2*10<sup>-3</sup> | ||
| + | |- | ||
| + | |Ampicillin | ||
| + | |10<sup>-3</sup> | ||
| + | |} | ||
| - | + | {| class="wikitable centre" width="25%" | |
| - | + | |+ 4 dishes with: | |
| - | - | + | |- |
| - | + | ! scope=col | Antibiotic | |
| - | + | ! scope=col | Concentration | |
| - | + | |- | |
| - | - | + | |Ampicillin |
| - | + | |4*10<sup>-4</sup> | |
| - | + | |} | |
| - | + | ||
| - | - | + | |
| - | + | ||
| - | + | ||
====Bacterial culture==== | ====Bacterial culture==== | ||
''by Laetitia'' | ''by Laetitia'' | ||
| - | After the bacterial transformation, we spread for each strain (LS or PS or GS) : | + | After the bacterial transformation, we spread for each strain (LS or PS or GS): |
| - | - | + | {| class="wikitable centre" width="25%" style="text-align:center" |
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | dish type | ||
| + | ! scope=col | 100µl | ||
| + | ! scope=col | 200µl | ||
| + | ! scope=col | concentrate | ||
| + | ! scope=col | control | ||
| + | |- | ||
| + | |Ampicillin | ||
| + | |X | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |Ampicillin+IPTG+XGal | ||
| + | |X | ||
| + | |X | ||
| + | |X | ||
| + | |X | ||
| + | |} | ||
| - | + | We leave the dishes overnight at 37°C. | |
| - | |||
| - | + | ==Photo of the Day== | |
| + | [[File:Paris Saclay 19_august.jpg|600px|center]] | ||
| - | + | '''Members present:''' | |
| + | * Instructors and advisors: Alice, Solenne and Sylvie. | ||
| + | * Students: Eugène, Hoang Vu, Laëtitia, Mélanie, Romain, Sean and Terry. | ||
{{Team:Paris_Saclay/notebook_footer}} | {{Team:Paris_Saclay/notebook_footer}} | ||
Latest revision as of 15:02, 14 October 2014
Contents |
Tuesday 19th August
Lab work
Chromoprotein + Lemon scent
Electrophoresis with the PpsI plasmide (obtain on thursday) + the chromoprotein PCR: we obtain results as expected
- ladder 10µl
- pPSI 1.2µl
- pPSI 1.2µl
- pPSI 1.2µl
- pPSI 1.2µl
- pPSI 1.2µl
- p cola 1.2µl
- PCR chromoprotein 2µl
Lemon Scent
Digestion of PCR results
by Sean, Hoang Vu, Eugene
| components | volumes |
|---|---|
| PCR purified LS | 15 μl |
| FastDigest green buffer 10X | 2 μl |
| PacI | 1 µl |
| H2O | 2 µl |
| components | volumes |
|---|---|
| PCR purified GS | 15 μl |
| FastDigest green buffer 10X | 2 μl |
| PacI | 1 µl |
| H2O | 2 µl |
| components | volumes |
|---|---|
| PCR purified PS | 15 μl |
| FastDigest green buffer 10X | 2 μl |
| PacI | 1 µl |
| H2O | 2 µl |
| components | volumes |
|---|---|
| Purified pPS1 | 30 μl |
| FastDigest green buffer 10X | 4 μl |
| PacI | 2 µl |
| H2O | 4 µl |
Purification of PCR products of limonen synthase
by Laetitia
We used the PCR products of limonen synthase prepared previously : August 18th
The five samples were pooled in one and then purified using this protocol : protocol
Bacterial transformation
by Laetitia
We used DH5alpha competent bacteria (prepared the 14/08) and they were transformed using the ligation product prepared previously August 18th using this protocol : protocol
We transformed 100µL of competent bacteria and we added the totality of the ligation product (pGEMTeasy containing LS or PS or GS)
Preparation of petri dishes
by Terry
| Antibiotic | Concentration |
|---|---|
| XGal | 80μg/ml |
| IPTG | 2*10-3 |
| Ampicillin | 10-3 |
| Antibiotic | Concentration |
|---|---|
| Ampicillin | 4*10-4 |
Bacterial culture
by Laetitia
After the bacterial transformation, we spread for each strain (LS or PS or GS):
| dish type | 100µl | 200µl | concentrate | control |
|---|---|---|---|---|
| Ampicillin | X | |||
| Ampicillin+IPTG+XGal | X | X | X | X |
We leave the dishes overnight at 37°C.
Photo of the Day
Members present:
- Instructors and advisors: Alice, Solenne and Sylvie.
- Students: Eugène, Hoang Vu, Laëtitia, Mélanie, Romain, Sean and Terry.
 "
"