|
|
| (114 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| - | {{Team:Paris_Saclay/default_header}} | + | {{Team:Paris_Saclay/modeling_header}} |
| - | | + | |
| - | | + | |
| - | | + | |
| | | | |
| | =Modeling= | | =Modeling= |
| | | | |
| | + | ==Overview== |
| | + | An important part of our [https://2014.igem.org/Team:Paris_Saclay/Project project] is devoted to the concrete realization of a lemon by moulding it from agar gel. It is well explained in the [https://2014.igem.org/Team:Paris_Saclay/Project/Lemon_Shaping lemon shaping] part. |
| | | | |
| | + | But, before starting more investigations and more experimentation, there is a real need to ensure that our project is feasible. We will tackle these following points : |
| | | | |
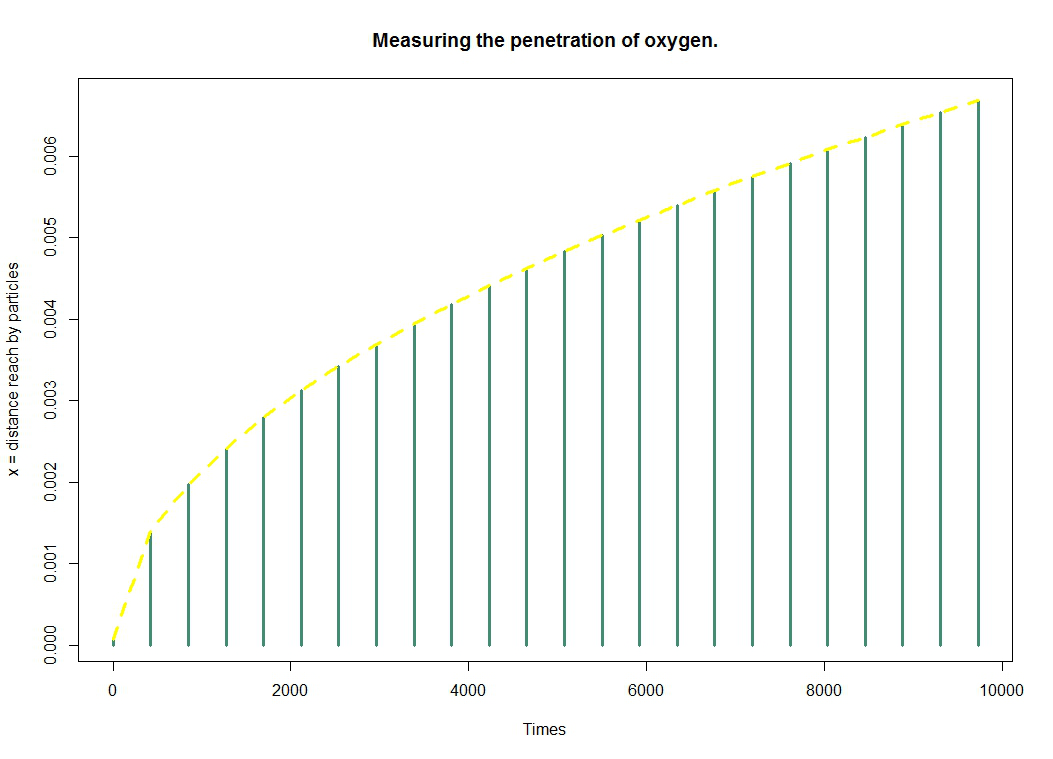
| | + | *By cutting a lemon, there is a slight yellow/green color on the surface and white on the inside. To reproduce this effect, we use a kill-switch system based on oxygen and had been achieved by the team iGEM Paris Saclay 2013. The more we penetrate into the gel, the less oxygen is present, the less kill-switch system is activated, the less bacteria will produce desired color, and the whiter the interior. For this, we must ensure that oxygen does not penetrate completely into the lemon. We decided to model [https://2014.igem.org/Team:Paris_Saclay/Modeling/oxygen_diffusion oxygen diffusion] into the gel. |
| | | | |
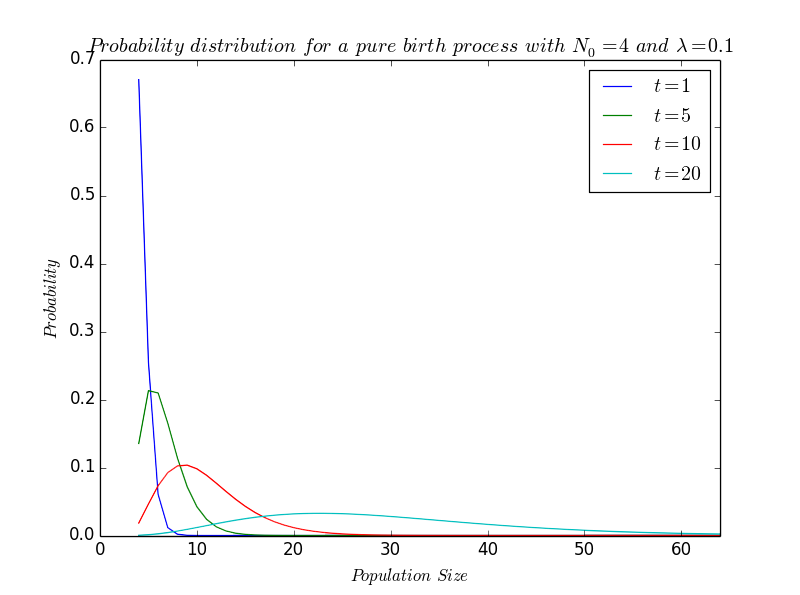
| | + | *A major issue is the development of our bacteria in an agar medium. We noticed that the bacteria inside the gel grew less than those on the outside. We model the bacterial growth |
| | | | |
| - | == Odor ==
| + | *To be even closer to the reality, we will imitate the [https://2014.igem.org/Team:Paris_Saclay/Project/Salicylate_Inducible_System ripening process] of a real lemon. We wanted to test if we could get a green color by fusion of two chromoproteins one yellow and one blue. |
| | + | |
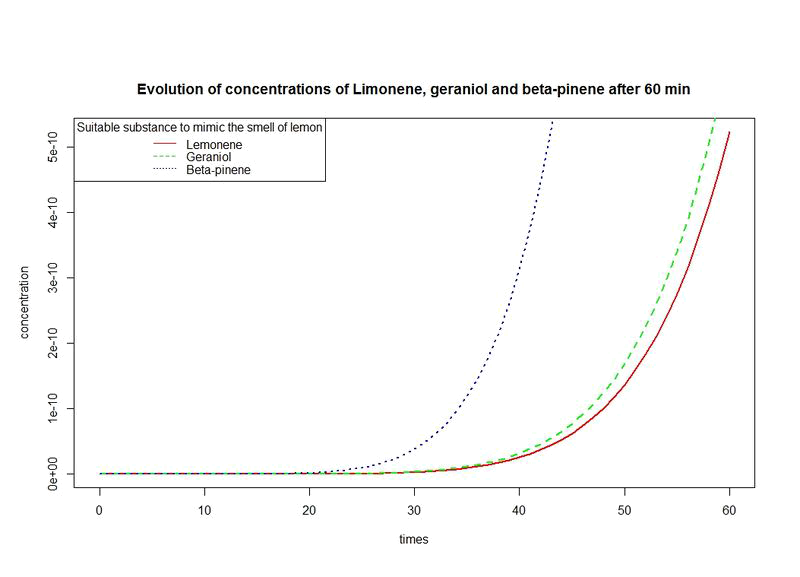
| | + | *Lastly, we obtain the [https://2014.igem.org/Team:Paris_Saclay/Project/Lemon_Scent lemon scent] by modifying ''E.coli'' in order to let it produce ''pinene'', ''limonene'' and ''geraniol'' but this does not ensure us that we will smell the lemon fragrance. |
| | | | |
| - | To synthesize the smell of lemon, we have to mainly produce Limonene, Geraniol and Pinene. The combination of these products will allow us to get the fragrance we need. We propose here a study of the evolution of these concentrations over time by using the technique Michaelis-Menten proposed in 1913 by Leonor Michaelis and Maud Menten. This will allow us to obtain a description of the kinetics of reactions between an enzyme (biological catalyst) and a substrate for giving a product.
| + | For all these reasons, modeling is really important for the concrete realization of our artwork and in general in an iGEM project! |
| | | | |
| - | === Basic model === | + | ==[https://2014.igem.org/Team:Paris_Saclay/Modeling/oxygen_diffusion Oxygen Diffusion]== |
| | | | |
| - | We consider the reations below between the enzyme $E$ and the substrate $S$. $ES$ is an complex foregoing the formation of the product P.
| |
| | | | |
| | + | [[File:Paris_Saclay_oxygenpenetration.png|left|150px]] |
| | | | |
| - | \[ E + S {\rightarrow}^{k_1} ES {\rightarrow}^{k_2} E + P \]
| + | ''Is it possible to have a crust on the surface of the lemon by producing a "lemon coloration-like" by our bacteria ?'' |
| - | \[ E + S {\leftarrow}_{k_{-1}} ES {\leftarrow}_{k_{-2}} E + P \]
| + | |
| | | | |
| - | where
| + | We invite you to discover how we modelled the [https://2014.igem.org/Team:Paris_Saclay/Modeling/oxygen_diffusion oxygen diffusion] in order to answer this question. |
| | | | |
| - | \[
| + | ==[https://2014.igem.org/Team:Paris_Saclay/Modeling/bacterial_Growth Bacterial Growth]== |
| - | \left\lbrace
| + | |
| - | \begin{array}{llll}
| + | |
| - | k_1 \text{ is the association constant of } E \text{ and } S \\
| + | |
| - | k_{-1} \text{ is the dissociation constant of } ES \\
| + | |
| - | k_2 \text{ is the reaction constant of } ES \text{ in } E + P \\
| + | |
| - | k_{-2}\text{ is the association constant of } E \text{ and } P \\
| + | |
| - | \end{array}
| + | |
| - | \right.
| + | |
| - | \]
| + | |
| | | | |
| - | We want to model the kinetics of the reaction catalyzed by enzymes Michaelis. We are interested in the concentration of molecules and various constants that we have described above.
| + | [[File:Paris_Saclay_Pure_Birth_Process.png|left|150px]] |
| | | | |
| - | The speed of the reaction $V$ is caracterised by the speed of apparition of the product. It is given by $V = \frac{d[P]}{dt}$ where $[P]$ is the concentration of the product.
| + | Now that we know if bacteria could develop in agar or not, a natural issue is to predict the [https://2014.igem.org/Team:Paris_Saclay/Modeling/bacterial_Growth bacterial growth]. |
| - | Modeling phenomena are inextricably linked to the reality of the phenomenon modeled, so we must take into account a number of experimental facts.
| + | So, this part aims at predicting over time the bacterial population growth on an ellipsoidal object - a fake lemon in practice -. |
| | + | More precisely, we have to determine the threshold of the initial proportion of bacteria for which we are sure that the population will never extinct... ''if this threshold exists!'' |
| | | | |
| | + | ==[https://2014.igem.org/Team:Paris_Saclay/Modeling/Fusion_Proteine Fusion Protein]== |
| | | | |
| - | * At the beginning, we don't have a product $P$. So the reaction $ES {\leftarrow}_{k_{-2}} E + P$ doesn't exist.
| + | [[File:Paris_Saclay_BlueChromoSwissModStruct.png|left|150px]] |
| - | * The speed of the reaction is constant equal to $V_0$. It only depend to the initiale concentration of substrate $[S]_0$ and enzyme $[E]_T$.
| + | |
| - | * Over time, the concentration of the substrate decrease while the concentration of P increase. We have only two possibles cases:
| + | |
| - |
| + | |
| - |
| + | |
| - | ** We have total reaction and it exist a time where $[S] = 0$. It imply that $V = 0$.
| + | |
| - | ** We reach a steady state between the formation and the destruction of the product. We have also $V = 0$.
| + | |
| - |
| + | |
| - | In fact we can consider that the speed of reaction decrease to 0. It will allow us to get an expression of $V_0$ in function of known parameters. But we must add others experimentals conditions. We consider the conditions set by Michaelis.
| + | |
| | | | |
| | + | An important aspect of our project is the color transition from green to yellow, in order to emulate the lemon maturation. |
| | + | To do this, we use a [https://2014.igem.org/Team:Paris_Saclay/Modeling/Fusion_Proteine fusion protein]. Before starting the fusion of the two chromoproteins -yellow and blue-, we have checked for their respective compatibility. That's why we discuss the structural modeling of the proteins. |
| | | | |
| - | ''' Michaelis condition '''
| + | ==[https://2014.igem.org/Team:Paris_Saclay/Modeling/odor Lemon Scent]== |
| | | | |
| - | To simplify the problem, Michaelis consider that:
| + | [[File:Paris_Saclay_odor_60.png|left|150px]] |
| | | | |
| | + | We need criteria to assess the scent of lemon. For this, we have process as follow: |
| | | | |
| - | * the concentration of substrate is very large relative to the concentration of enzyme: $[S] >> [E]_T $ and the concentration of the product is negligible: $P \approx 0$. With the equation $ES {\leftarrow}_{k_{-2}} E + P$, we know that the speed formation of $ES$ is gived by $v = k_{-2}[E][P] \approx 0$.
| + | * We evaluate the evolution of the concentrations of three fragrances over time. |
| - | | + | * We give a procedure for estimating a suitable time interval in which we can say that we got the lemon smell as desired. |
| - | * We suppose that we reach the equilibrium between $[E], [S]$ and $[ES]$ fastly. In the case of equilibrium, $[ES]$ is constant until $[P] \approx 0$, and so | + | |
| - | $\frac{d[ES]}{dt} = 0$.
| + | |
| - | | + | |
| - | * $[S]_0 >> [E]_T \rightarrow [ES]_{max} << [S]_0$ because the concentration of the complex $[ES]$ is limited by the concentration of enzyme.
| + | |
| - | Since $[S] = [S]_0 - [ES]$ and $[ES] \approx 0$, we have $[S] = [S]_0$.
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | In the following, we will stay in the conditions of Michaelis.
| + | |
| - | | + | |
| - | $\forall t, V = \frac{d[P]}{dt} = k_2 [ES] - k_{-2}[E][P]$. Since initially $[P] \approx 0$, we have $\boxed{V_i = k_2 [ES]}$. The problem is that $[ES]$ is unknow ! \newline
| + | |
| - | | + | |
| - | The variation of the concentration of the complexe $ES$ is given by the difference between it formation and it dissociation. This imply
| + | |
| - | | + | |
| - | \begin{align*}
| + | |
| - | %
| + | |
| - | & \frac{d[ES]}{dt} = k_1 [E][S ] - (k_{-1} + k_2) [ES] = 0 \text{ in initial condition }. \\
| + | |
| - | & \text{ So, } \frac{[E][S]}{[ES]} = \frac{k_{-1} + k_2}{k1} =: Km = \text{ Michaelis constant }. \\
| + | |
| - | & \text{ We deduce the Michaelis equation } \boxed{[ES] = \frac{[E][S]}{Km}}.
| + | |
| - | %
| + | |
| - | \end{align*}
| + | |
| - | | + | |
| - | For all time, the total enzyme $E$ is composed by free molecule of $E$ and molecule associated to the substrate to form $ES$. It is expressed by
| + | |
| - | $\forall t, [E]_T = [E] + [ES]$, where we deduce $[E] = [E]_T - [ES]$. Replacing this in the Michaelis equation, we have :
| + | |
| - | | + | |
| - | \begin{align*}
| + | |
| - | %
| + | |
| - | & [ES] = \frac{([E]_T - [ES])[S]}{Km} = \frac{[E]_T [S]}{Km} - \frac{[ES][S]}{Km}. \\
| + | |
| - | & [ES](1 + \frac{[S]}{Km}) = \frac{[E]_T [S]}{Km} \\
| + | |
| - | & [ES] = \frac{\frac{[E]_T [S]}{Km} }{ \frac{Km + [S]}{Km} } = [E]_T \times \frac{[S]}{Km + [S]}.
| + | |
| - | %
| + | |
| - | \end{align*}
| + | |
| - | | + | |
| - | Returning to the expression of $V_i = k_2 [ES]$ and replacing $[ES]$ by the last expression below, we have $V_i = k_2 [E]_T \times \frac{[S]}{Km + [S]} $
| + | |
| - | | + | |
| - | \[ \text{Since }
| + | |
| - | \left\lbrace
| + | |
| - | \begin{array}{ll}
| + | |
| - | [ES] \leq [E]_T \\
| + | |
| - | V_i = k_2 [ES]
| + | |
| - | \end{array}
| + | |
| - | \right.
| + | |
| - | \text{ we have } \boxed{ V_{max} = k_2 [E]_T } (k_2 \text{ is also noted } k_{cat}).
| + | |
| - | \]
| + | |
| - | | + | |
| - | Taking into account all these results, we obtain the following equation of Michaelis-Menten
| + | |
| - | | + | |
| - | \[ \boxed{ V_i = V_{max} \frac{[S]}{[S] + Km} } . \]
| + | |
| - | | + | |
| - | We can determine $V_{max}$ and $Km$ experimentally. $[S] \approx [S]_0$ is the concentration of substrate at the beginning.
| + | |
| - | | + | |
| - | | + | |
| - | In the following, we will work directly with this expression.
| + | |
| - | | + | |
| - | | + | |
| - | === Biosynthetic pathway of limonene ===
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | === Complete model ===
| + | |
| - | | + | |
| - | By analysing the different reaction in pathway above and using the equation of Michaelis-Menten, we obtain the system of nine equation below:
| + | |
| - | | + | |
| - | | + | |
| - | \[
| + | |
| - | \left\lbrace
| + | |
| - | \begin{array}{llllllllll}
| + | |
| - | \frac{d[acetyl-CoA]}{dt} &= - \frac{Vm1 [acetyl-CoA]}{Km1 + [acetyl-CoA]} \\
| + | |
| - | \frac{d[acetoacetyl-CoA]}{dt} &= \frac{Vm1 [acetyl-CoA]}{Km1 + [acetyl-CoA]} - \frac{Vm2 [acetoacetyl-CoA]}{Km2 + [acetoacetyl-CoA]} \\
| + | |
| - | \frac{d[HMG-CoA]}{dt} &= \frac{Vm2 [acetoacetyl-CoA]}{Km2 + [acetoacetyl-CoA]} - \frac{Vm3 [HMG-CoA]}{Km3 + [HMG-CoA]} \\
| + | |
| - | \frac{d[Mevalonate]}{dt} &= \frac{Vm3 [HMG-CoA]}{Km3 + [HMG-CoA]} - \frac{Vm4 [Mevalonate]}{Km4 + [Mevalonate]} \\
| + | |
| - | \frac{d[MevalonateP-5]}{dt} &= \frac{Vm4 [Mevalonate]}{Km4 + [Mevalonate]} - \frac{Vm5 [MevalonateP-5]}{Km5 + [MevalonateP-5]} \\
| + | |
| - | \frac{d[5-pyroP-MevalonateP]}{dt} &= \frac{Vm5 [MevalonateP-5]}{Km5 + [MevalonateP-5]} - \frac{Vm6 [5-pyroP-MevalonateP]}{Km6 + [5-pyroP-MevalonateP]} \\
| + | |
| - | \frac{d[IPP]}{dt} &= \frac{Vm6 \frac{[5-pyroP-MevalonateP]}{2}}{Km6 + \frac{[5-pyroP-MevalonateP]}{2}} - \frac{Vm7 \frac{[IPP]}{2}}{Km7 + \frac{[IPP]}{2}} - \frac{Vm8 \frac{[IPP]}{2}}{Km8 + \frac{[IPP]}{2}} \\
| + | |
| - | \frac{d[DMAPP]}{dt} &= \frac{Vm6 \frac{[5-pyroP-MevalonateP]}{2}}{Km6 + \frac{[5-pyroP-MevalonateP]}{2}} + \frac{Vm7 \frac{[IPP]}{2}}{Km7 + \frac{[IPP]}{2}} - \frac{Vm8 \frac{[DMAPP]}{2}}{Km8 + \frac{[DMAPP]}{2}} \\
| + | |
| - | \frac{d[GPP]}{dt} &= \frac{Vm7 \frac{[IPP]}{2}}{Km7 + \frac{[IPP]}{2}} + \frac{Vm8 \frac{[DMAPP]}{2}}{Km8 + \frac{[DMAPP]}{2}} - \frac{Vm_f [GPP]}{Km_f + [GPP]} \\
| + | |
| - | \frac{d[Fragrance]}{dt} &= \frac{Vm_f [GPP]}{Kmf + [GPP]}
| + | |
| - | \end{array}
| + | |
| - | \right.
| + | |
| - | \]
| + | |
| - | | + | |
| - | where $Vm_f$ and $Km_f$ are constant relative to the Fragrance ie Limonene, Geraniol or Pinene.
| + | |
| - | | + | |
| - | To solve these equation, we use the logiciel R and we obtain these following results for the production of Limonene:
| + | |
| - | | + | |
| - | The differents constant are obtained via Brenda enzyme database: http://www.brenda-enzymes.org/
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | == Oxygen Diffusion ==
| + | |
| - | | + | |
| - | To realise our artwork, we use an agarose gel to obtain the shape of a lemon. To push the resemblance to the extreme, we wish to have a crust in the edge of the lemon when we seperate it. In fact, we build bacteria who produce yellow/green color in presence of oxygen. Thus we must evaluate the penetration of the oxygen in the gel !
| + | |
| - | | + | |
| - | Oxygen penetrates into the gel by diffusion phenomenon that we will study below. We first use the following phenomenological law suggest by Adolphe Fick in 1855:
| + | |
| | | | |
| | | | |
| | ---- | | ---- |
| - |
| |
| - | '' In an homogeneous and isotropic environment, containing particles distributed inhomogeneously,appears spontaneously a volumetric flow density vector particle $\overrightarrow{J}(M,t) $. In any point $M$ in space, this vector is proportional to the gradient of the particle density $n(M,t)$. Mathematicaly, this relationship take the form: \[ \overrightarrow{J}(M,t) = - D \times \nabla n(M,t) \qquad (1) \] where $D$ is the diffusion coefficient and $\nabla$ is the gradient vector.''
| |
| - |
| |
| - | ----
| |
| - |
| |
| - | In equation after Fick's law above, we need to know the parameter $D$. We then searched in scientific literature and article '''[1]''' describes a method to get it. Referring to this article, the diffusion coefficient of oxygen in agarose is $ D = 0{,}256 \times 10^{-8} m^2 s^{-1} $.
| |
| - |
| |
| - | The phenomenon we are facing is quite difficult to tackle if taken as a whole and We must emit a number of intuitively acceptable hypothesis.
| |
| - |
| |
| - | * To simplify the problem, we consider that the diffusion of oxygen particle occurs only in one direction. So $\overrightarrow{J}(M,t) = J(x,t) \overrightarrow{e}_x $. This hypothesis is credible because we seek the maximum penetrance of oxygen. This is why we consider that the diffusion occurs in the line of greatest slope.
| |
| - |
| |
| - | * Spatial variations in the density of particles are connected to spatial variations of the vector $\overrightarrow{J}(M,t)$ by '''the material's equation of conservation''' in presence of volume distribution of particle source $\sigma (x,t)$ (device which injects or subtracted particles to the system) :
| |
| - | \[ \frac{\partial n}{\partial t} (x,t) = - \frac{\partial J}{\partial x} (x,t) + \sigma (x,t) \qquad (2) \]
| |
| - |
| |
| - | By replacing $(2)$ in $(1)$, we obtain the following '''equation of diffusion''' :
| |
| - | \[ \forall t, \forall x, \bigg( \frac{\partial}{\partial t} - D \frac{\partial^2}{\partial x^2} \bigg) n(x,t) = \sigma (x,t) \qquad (3) .\]
| |
| - |
| |
| - | As our lemon is exposed to the ambient air, we stay in steady state where the source $ \sigma (x,t) $ is equal to $N_0$ the quantity of $O_2$ in the air.
| |
| - |
| |
| - | To solve this equation, we use Fourier's analysis and it is kwown (classical solution of the heat equation) that
| |
| - |
| |
| - | \[ \forall x, \forall t>0, n(x,t) = \frac{N_0}{\sqrt{4 \pi D t}} exp \bigg(- \frac{x^2}{4 D t} \bigg) + \int_{0}^{t} \underbrace{N_0 * exp \bigg(- \frac{|x|^2}{4 D \tau} \bigg)}_{= 0 \text{ by symmetry of the gaussian distribution }} \frac{d\tau}{\sqrt{4 \pi D \tau} } \]
| |
| - |
| |
| - | Whence, \[ \forall x, \forall t>0, n(x,t) = \frac{N_0}{\sqrt{4 \pi D t}} exp \bigg(- \frac{x^2}{4 D t} \bigg) \]
| |
| - |
| |
| - | Thus we deduce easily that the average dispersion particle is given by the variance $\Delta x = \sqrt{2Dt}$. Using this formula, we deduct that, for example, oxygen will penetrate $3 \times 10^{-3} m$ in $1956.522 s = 32.6082 $ minutes.
| |
| - |
| |
| - | The charts below give us a very clear idea of penetrance evaluated over time.
| |
| - |
| |
| - | === Graphical visualization ===
| |
| - |
| |
| - | [[File:Paris_Saclay_oxygenGraph.jpeg|600px]]
| |
| - |
| |
| - |
| |
| - |
| |
| - |
| |
| - |
| |
| - | [[References:]]
| |
| - |
| |
| - | '''[1]''' A.C. Hulst, H.J.H. Hens, R.M. Buitelaar and J. Tramper, ''Determination of the effective diffusion coefficient of oxygen in gel materials in relation to gel concentration'', Biotechnology Techniques Vol 3 No 3 199-204 (1989).
| |
| - |
| |
| - | '''[2]''' Vincent Renvoizé, ''Physique PC-PC*'', Cap Prepas, Pearson Education, 2010.
| |
| - |
| |
| - | '''[3]''' Gilles Camus, La cinétique des enzymes michaeliennes et l'équation de Michaelis-Menten, 22 novembre 2012.
| |
| - |
| |
| - |
| |
| - |
| |
| | | | |
| | | | |
| | | | |
| | + | '''Note''' : Do not be afraid and run away because of seing mathematical formulae : ''Maths is fun'' and we have made a real effort to be as precise as possible in order non-mathematicians to understand. For example, in order not to disgust poor mathematical readers of others, we will just add one or two -for the warrior- star(s) to parts which need more mathematical background. Morever, we illustrated our work with figures which could be understood for themselves, without knowing the method to get them. |
| | | | |
| | {{Team:Paris_Saclay/default_footer}} | | {{Team:Paris_Saclay/default_footer}} |

Modeling
Overview
An important part of our project is devoted to the concrete realization of a lemon by moulding it from agar gel. It is well explained in the lemon shaping part.
But, before starting more investigations and more experimentation, there is a real need to ensure that our project is feasible. We will tackle these following points :
- By cutting a lemon, there is a slight yellow/green color on the surface and white on the inside. To reproduce this effect, we use a kill-switch system based on oxygen and had been achieved by the team iGEM Paris Saclay 2013. The more we penetrate into the gel, the less oxygen is present, the less kill-switch system is activated, the less bacteria will produce desired color, and the whiter the interior. For this, we must ensure that oxygen does not penetrate completely into the lemon. We decided to model oxygen diffusion into the gel.
- A major issue is the development of our bacteria in an agar medium. We noticed that the bacteria inside the gel grew less than those on the outside. We model the bacterial growth
- To be even closer to the reality, we will imitate the ripening process of a real lemon. We wanted to test if we could get a green color by fusion of two chromoproteins one yellow and one blue.
- Lastly, we obtain the lemon scent by modifying E.coli in order to let it produce pinene, limonene and geraniol but this does not ensure us that we will smell the lemon fragrance.
For all these reasons, modeling is really important for the concrete realization of our artwork and in general in an iGEM project!
Is it possible to have a crust on the surface of the lemon by producing a "lemon coloration-like" by our bacteria ?
We invite you to discover how we modelled the oxygen diffusion in order to answer this question.
Now that we know if bacteria could develop in agar or not, a natural issue is to predict the bacterial growth.
So, this part aims at predicting over time the bacterial population growth on an ellipsoidal object - a fake lemon in practice -.
More precisely, we have to determine the threshold of the initial proportion of bacteria for which we are sure that the population will never extinct... if this threshold exists!
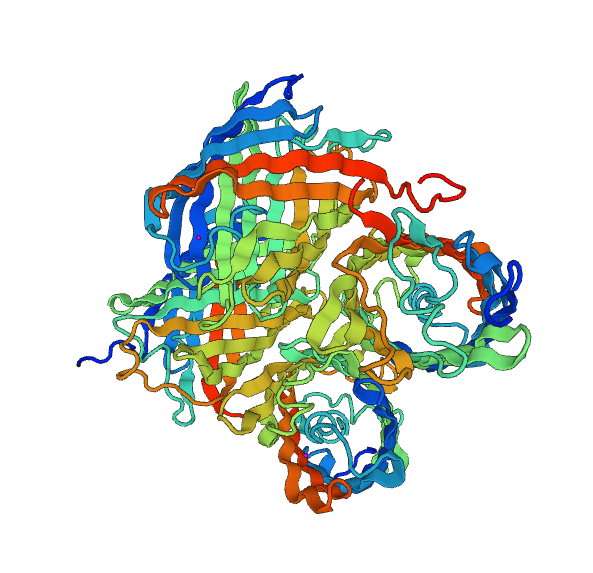
An important aspect of our project is the color transition from green to yellow, in order to emulate the lemon maturation.
To do this, we use a fusion protein. Before starting the fusion of the two chromoproteins -yellow and blue-, we have checked for their respective compatibility. That's why we discuss the structural modeling of the proteins.
We need criteria to assess the scent of lemon. For this, we have process as follow:
- We evaluate the evolution of the concentrations of three fragrances over time.
- We give a procedure for estimating a suitable time interval in which we can say that we got the lemon smell as desired.
Note : Do not be afraid and run away because of seing mathematical formulae : Maths is fun and we have made a real effort to be as precise as possible in order non-mathematicians to understand. For example, in order not to disgust poor mathematical readers of others, we will just add one or two -for the warrior- star(s) to parts which need more mathematical background. Morever, we illustrated our work with figures which could be understood for themselves, without knowing the method to get them.

 "
"