Team:KIT-Kyoto/Notebook/Labnote
From 2014.igem.org
| (110 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
{{Team:KIT-Kyoto/slidemenu}} | {{Team:KIT-Kyoto/slidemenu}} | ||
{{Team:KIT-Kyoto/navi}} | {{Team:KIT-Kyoto/navi}} | ||
| + | {{Team:KIT-Kyoto/close_navi}} | ||
{{Team:KIT-Kyoto/table}} | {{Team:KIT-Kyoto/table}} | ||
{{Team:KIT-Kyoto/Scroll}} | {{Team:KIT-Kyoto/Scroll}} | ||
| Line 11: | Line 12: | ||
<!--サイドメニュー--> | <!--サイドメニュー--> | ||
<ul id="nav5"> | <ul id="nav5"> | ||
| - | <li class="menuimg"><a href="/Team:KIT-Kyoto | + | <li class="menuimg"><a href="/Team:KIT-Kyoto">home</a></li> |
| - | <li class="menuimg"><a href="/Team:KIT-Kyoto | + | <li class="menuimg"><a href="/Team:KIT-Kyoto/About">about us</a> |
| - | <li class="menuimg"><a href="/Team:KIT-Kyoto | + | <li class="menuimg"><a href="/Team:KIT-Kyoto/Project">project</a></li> |
| + | <li class="menuimg"><a href="/Team:KIT-Kyoto/HumanPractice">policy & practices</a></li> | ||
| + | <li class="menuimg"><a href="/Team:KIT-Kyoto/Achievement">Achievement</a></li> | ||
</ul> | </ul> | ||
<div class="active"> | <div class="active"> | ||
| Line 19: | Line 22: | ||
</div> | </div> | ||
<ul class="submenu"> | <ul class="submenu"> | ||
| - | <li class="category"><a href="javascript:void(0) | + | <li class="category"> |
| - | <ul class="slidemenu"> | + | <a href="javascript:void(0)"> |
| - | <li class="btn"><a href=" | + | <font color="#143">Click Here To Enlarge</font> LabNote |
| - | <li class="btn"><a href=" | + | </a> |
| - | <li class="btn"><a href=" | + | </li> |
| - | <li class="btn"><a href=" | + | <ul class="slidemenu"> |
| - | + | <li class="btn"> | |
| - | + | <a href="">June | |
| - | + | </a> | |
| - | <li class="btn" | + | </li> |
| - | + | <li class="btn"> | |
| - | + | <a href="">July | |
| - | + | </a> | |
| - | + | </li> | |
| - | + | <li class="btn"> | |
| - | + | <a href="">August | |
| - | + | </a> | |
| - | + | </li> | |
| + | <li class="btn"> | ||
| + | <a href="">September | ||
| + | </a> | ||
| + | </li> | ||
| + | <li class="btn"> | ||
| + | <a href="">October | ||
| + | </a> | ||
| + | </li> | ||
| + | </ul> | ||
| + | <li><a href="/Team:KIT-Kyoto/Notebook/Protocol">Protocol</a></li> | ||
</ul> | </ul> | ||
</div> | </div> | ||
| Line 43: | Line 56: | ||
<div id="container"> | <div id="container"> | ||
<div class="main-contents"> | <div class="main-contents"> | ||
| + | <div id="ln"> | ||
| + | <div class="accordion"> | ||
<h1>LabNote</h1> | <h1>LabNote</h1> | ||
| - | <h2>June</h2> | + | <h2><a href="javascript:void(0);">June</a></h2> |
<div class="scroll"></div> | <div class="scroll"></div> | ||
| + | <div class="ac"> | ||
<!-- タイトル --> | <!-- タイトル --> | ||
<h3>June 10, 2014</h3> | <h3>June 10, 2014</h3> | ||
| Line 57: | Line 73: | ||
<!--Miniprep--> | <!--Miniprep--> | ||
<strong>Miniprep</strong> | <strong>Miniprep</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 86: | Line 103: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Miniprep終わり--> | <!--Miniprep終わり--> | ||
| Line 92: | Line 110: | ||
<!--Restriction Digest--> | <!--Restriction Digest--> | ||
<strong>Restriction Digest</strong> | <strong>Restriction Digest</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 104: | Line 123: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td>BL21(DE3) | + | <td>BL21(DE3)</td> |
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td>BL21(DE3) | + | <td>BL21(DE3)</td> |
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td>BL21(DE3) | + | <td>BL21(DE3)</td> |
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td>BL21(DE3) | + | <td>BL21(DE3)</td> |
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
| - | <td> | + | <td>CuMTS62</td> |
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td>BL21(DE3) | + | <td>BL21(DE3)</td> |
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td><em>Sal</em>1, <em>Not</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td>BL21(DE3) | + | <td>BL21(DE3)</td> |
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td><em>Sal</em>1, <em>Not</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td>BL21(DE3) | + | <td>BL21(DE3)</td> |
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td><em>Sal</em>1, <em>Not</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td>BL21(DE3) | + | <td>BL21(DE3)</td> |
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
| - | <td> | + | <td>CuMTS62</td> |
| - | <td> | + | <td><em>Sal</em>1, <em>Not</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Ristriction Digest--> | <!--Ristriction Digest--> | ||
<br> | <br> | ||
| Line 165: | Line 185: | ||
</p> | </p> | ||
<!--本文終わり--> | <!--本文終わり--> | ||
| - | |||
<!-- タイトル --> | <!-- タイトル --> | ||
<h3>June 11, 2014</h3> | <h3>June 11, 2014</h3> | ||
| Line 177: | Line 196: | ||
<!--AGE--> | <!--AGE--> | ||
<strong>AGE</strong> | <strong>AGE</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
<th rowspan="2">Lane</th> | <th rowspan="2">Lane</th> | ||
| - | <th colspan=" | + | <th colspan="3">Sample</th> |
<th rowspan="2">Enzyme</th> | <th rowspan="2">Enzyme</th> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| + | <th>Marker</th> | ||
<th>Vector</th> | <th>Vector</th> | ||
<th>Insert</th> | <th>Insert</th> | ||
| Line 190: | Line 211: | ||
<td>1</td> | <td>1</td> | ||
<td>λDNA-HindⅢ</td> | <td>λDNA-HindⅢ</td> | ||
| - | <td | + | <td></td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>2</td> | <td>2</td> | ||
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>Non Treated</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>3</td> | <td>3</td> | ||
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>4</td> | <td>4</td> | ||
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td><em>Sal</em>1, <em>Not</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>5</td> | <td>5</td> | ||
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>Non Treated</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>6</td> | <td>6</td> | ||
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>7</td> | <td>7</td> | ||
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td><em>Sal</em>1, <em>Not</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>8</td> | <td>8</td> | ||
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>Non Treated</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>9</td> | <td>9</td> | ||
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>10</td> | <td>10</td> | ||
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
| - | <td> | + | <td>CuMTS3</td> |
| - | <td> | + | <td><em>Sal</em>1, <em>Not</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>11</td> | <td>11</td> | ||
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>Non Treated</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>12</td> | <td>12</td> | ||
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
| + | </tr> | ||
| + | <tr> | ||
| + | <td>13</td> | ||
| + | <td></td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>CuMTS62</td> | ||
| + | <td><em>Sal</em>1, <em>Not</em>1</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--AGE--> | <!--AGE--> | ||
| + | <img src="/wiki/images/8/88/Kit_June11_c.jpg"> | ||
<br> | <br> | ||
<br> | <br> | ||
| Line 276: | Line 319: | ||
<!--Transformation--> | <!--Transformation--> | ||
<strong>Transformation</strong> | <strong>Transformation</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 305: | Line 349: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Transformation終わり--> | <!--Transformation終わり--> | ||
<br> | <br> | ||
| Line 319: | Line 364: | ||
<!--Colony Isolation--> | <!--Colony Isolation--> | ||
<strong>Colony Isolation</strong><br> | <strong>Colony Isolation</strong><br> | ||
| - | Select 6 colonies per plate from 4 plates from June | + | Select 6 colonies per plate from 4 plates from June 12.<br> |
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 351: | Line 397: | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
| - | <td> | + | <td>CuMTS62</td> |
<td>6-a, 6-b</td> | <td>6-a, 6-b</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Colony Isolation--> | <!--Colony Isolation--> | ||
<br> | <br> | ||
| Line 374: | Line 421: | ||
<!--Miniprep--> | <!--Miniprep--> | ||
<strong>Miniprep</strong> | <strong>Miniprep</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 423: | Line 471: | ||
</table> | </table> | ||
| + | </div> | ||
<!--Miniprep終わり--> | <!--Miniprep終わり--> | ||
<br> | <br> | ||
<!--Miniprep--> | <!--Miniprep--> | ||
<strong>Miniprep</strong> | <strong>Miniprep</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 457: | Line 507: | ||
</table> | </table> | ||
| + | </div> | ||
<!--Miniprep終わり--> | <!--Miniprep終わり--> | ||
<br> | <br> | ||
<!--Restriction Digest--> | <!--Restriction Digest--> | ||
<strong>Restriction Digest</strong> | <strong>Restriction Digest</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 476: | Line 528: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 483: | Line 535: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 490: | Line 542: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 497: | Line 549: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 504: | Line 556: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 511: | Line 563: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 518: | Line 570: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 525: | Line 577: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 532: | Line 584: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 539: | Line 591: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 546: | Line 598: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 553: | Line 605: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Ristriction Digest--> | <!--Ristriction Digest--> | ||
| Line 562: | Line 615: | ||
<!--AGE--> | <!--AGE--> | ||
<strong>AGE</strong> | <strong>AGE</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
<th rowspan="2">Lane</th> | <th rowspan="2">Lane</th> | ||
| - | <th colspan=" | + | <th colspan="3">Sample</th> |
<th rowspan="2">Insert</th> | <th rowspan="2">Insert</th> | ||
<th rowspan="2">Enzyme</th> | <th rowspan="2">Enzyme</th> | ||
| Line 571: | Line 625: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| + | <th>Marker</th> | ||
<th>Host</th> | <th>Host</th> | ||
<th>Vector</th> | <th>Vector</th> | ||
| Line 577: | Line 632: | ||
<td>1</td> | <td>1</td> | ||
<td>1kb Ladder</td> | <td>1kb Ladder</td> | ||
| - | <td | + | <td></td> |
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
<tr> | <tr> | ||
<td>2</td> | <td>2</td> | ||
<td>λDNA-HindⅢ</td> | <td>λDNA-HindⅢ</td> | ||
| - | <td | + | <td></td> |
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
<tr> | <tr> | ||
<td>3</td> | <td>3</td> | ||
| + | <td></td> | ||
<td>1-a</td> | <td>1-a</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>4</td> | <td>4</td> | ||
| + | <td></td> | ||
<td>1-b</td> | <td>1-b</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>5</td> | <td>5</td> | ||
| + | <td></td> | ||
<td>2-a</td> | <td>2-a</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>6</td> | <td>6</td> | ||
| + | <td></td> | ||
<td>2-b</td> | <td>2-b</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>7</td> | <td>7</td> | ||
| + | <td></td> | ||
<td>3-a</td> | <td>3-a</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>8</td> | <td>8</td> | ||
| + | <td></td> | ||
<td>3-b</td> | <td>3-b</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<td>9</td> | <td>9</td> | ||
| + | <td></td> | ||
<td>6-a</td> | <td>6-a</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
<tr> | <tr> | ||
<td>10</td> | <td>10</td> | ||
| + | <td></td> | ||
<td>6-b</td> | <td>6-b</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>11</td> | <td>11</td> | ||
| + | <td></td> | ||
<td>BL21(DE3)</td> | <td>BL21(DE3)</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>12</td> | <td>12</td> | ||
| + | <td></td> | ||
<td>BL21(DE3)</td> | <td>BL21(DE3)</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>13</td> | <td>13</td> | ||
| + | <td></td> | ||
<td>BL21(DE3)</td> | <td>BL21(DE3)</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>14</td> | <td>14</td> | ||
| + | <td></td> | ||
<td>BL21(DE3)</td> | <td>BL21(DE3)</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--AGE--> | <!--AGE--> | ||
<br> | <br> | ||
| - | + | Apply λDNA-HindⅢ to compare and contrast AGE of July 14 and June 11. | |
<br> | <br> | ||
| + | <img src="/wiki/images/b/b6/Kit_June14_c.jpg"> | ||
<br> | <br> | ||
</p> | </p> | ||
| Line 696: | Line 773: | ||
<!--Preculture--> | <!--Preculture--> | ||
<strong>Preculture</strong> | <strong>Preculture</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 726: | Line 804: | ||
<tr> | <tr> | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
| - | <td | + | <td>pGEX6P-2</td> |
| + | <td></td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Preculture終わり--> | <!--Preculture終わり--> | ||
<br> | <br> | ||
<!--Main Culture--> | <!--Main Culture--> | ||
<strong>Main Culture</strong> | <strong>Main Culture</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 745: | Line 826: | ||
<tr> | <tr> | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
| - | <td | + | <td>pGEX6P-2</td> |
| + | <td>CuMTSE1</td> | ||
<td>Induced</td> | <td>Induced</td> | ||
</tr> | </tr> | ||
| + | <tr> | ||
| + | <td>BL21(DE3)pLysS</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>CuMTSE2</td> | ||
| + | <td>Induced</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BL21(DE3)pLysS</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>CuMTS3</td> | ||
| + | <td>Induced</td> | ||
| + | <tr> | ||
| + | <td>BL21(DE3)pLysS</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>CuMTS62</td> | ||
| + | <td>Induced</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BL21(DE3)pLysS</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td></td> | ||
| + | <td>Induced</td> | ||
<tr> | <tr> | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| + | <td>Non-induced</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BL21(DE3)pLysS</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>CuMTSE2</td> | ||
| + | <td>Non-induced</td> | ||
| + | </tr><tr> | ||
| + | <td>BL21(DE3)pLysS</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>CuMTS3</td> | ||
| + | <td>Non-induced</td> | ||
| + | </tr><tr> | ||
| + | <td>BL21(DE3)pLysS</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>CuMTS62</td> | ||
| + | <td>Non-induced</td> | ||
| + | </tr><tr> | ||
| + | <td>BL21(DE3)pLysS</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td></td> | ||
| + | <td>Non-induced</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <!--Main Culture終わり--> | ||
| + | <br> | ||
| + | </p> | ||
| + | <!--本文終わり--> | ||
| + | <br> | ||
| + | <!-- タイトル --> | ||
| + | <h3>June 18, 2014</h3> | ||
| + | <!-- タイトル終わり --> | ||
| + | |||
| + | <!-- 本文開始 --> | ||
| + | <p class="sentence"> | ||
| + | <!-- 実験者名 --> | ||
| + | YAMAJI<br> | ||
| + | <!-- 実験者名終わり --> | ||
| + | <!--Preculture--> | ||
| + | <strong>Preculture</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Host</th> | ||
| + | <th colspan="2">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BL21(DE3)pLysS</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>CuMTSE1</td> | ||
| + | </tr> | ||
| + | <td>BL21(DE3)pLysS</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>CuMTSE2</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <!--Preculture終わり--> | ||
| + | <br> | ||
| + | <!--Main Culture--> | ||
| + | <strong>Main Culture</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Host</th> | ||
| + | <th colspan="2">Sample</th> | ||
| + | <th rowspan="2">IPTG</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BL21(DE3)pLysS</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td></td> | ||
<td>Induced</td> | <td>Induced</td> | ||
</tr> | </tr> | ||
| Line 757: | Line 942: | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
| - | <td> | + | <td>CuMTSE1</td> |
<td>Induced</td> | <td>Induced</td> | ||
| + | </tr> | ||
<tr> | <tr> | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
| - | <td | + | <td>pGEX6P-2</td> |
| - | <td>Non- | + | <td>CuMTSE2</td> |
| + | <td>Induced</td> | ||
| + | <tr> | ||
| + | <td>BL21(DE3)pLysS</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td></td> | ||
| + | <td>Non-induced</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
| - | <td> | + | <td>CuMTSE1</td> |
| - | <td>Non- | + | <td>Non-induced</td> |
<tr> | <tr> | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td>Non- | + | <td>Non-induced</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Main Culture終わり--> | <!--Main Culture終わり--> | ||
<br> | <br> | ||
| - | <!--Protein Extraction (E.coli)--> | + | <!--Protein Extraction (<em>E. coli</em>)--> |
| - | <strong>Protein Extraction (E.coli)</strong> | + | <strong>Protein Extraction (<em>E. coli</em>)</strong> |
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 792: | Line 986: | ||
<tr> | <tr> | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
| - | <td | + | <td>pGEX6P-2</td> |
| + | <td></td> | ||
<td>Induced</td> | <td>Induced</td> | ||
</tr> | </tr> | ||
| Line 804: | Line 999: | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
| - | <td> | + | <td>CuMTSE2</td> |
<td>Induced</td> | <td>Induced</td> | ||
<tr> | <tr> | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
| - | <td | + | <td>pGEX6P-2</td> |
| - | <td>Non- | + | <td></td> |
| + | <td>Non-induced</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
| - | <td> | + | <td>CuMTSE1</td> |
| - | <td>Non- | + | <td>Non-induced</td> |
<tr> | <tr> | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td>Non- | + | <td>Non-induced</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| - | <!--Protein Extraction (E.coli)終わり--> | + | </div> |
| + | <!--Protein Extraction (<em>E. coli</em>)終わり--> | ||
<br> | <br> | ||
<!--Preparations for SDS-PAGE--> | <!--Preparations for SDS-PAGE--> | ||
| Line 831: | Line 1,028: | ||
<!--Preparations for SDS-PAGE終わり--> | <!--Preparations for SDS-PAGE終わり--> | ||
<br> | <br> | ||
| + | <img src="/wiki/images/0/09/Kit_June18_c.jpg"> | ||
<br> | <br> | ||
<br> | <br> | ||
| Line 847: | Line 1,045: | ||
<!--SDS-PAGE--> | <!--SDS-PAGE--> | ||
<strong>SDS-PAGE</strong> | <strong>SDS-PAGE</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 859: | Line 1,058: | ||
<tr> | <tr> | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
| - | <td | + | <td>pGEX6P-2</td> |
| + | <td></td> | ||
<td>Induced</td> | <td>Induced</td> | ||
</tr> | </tr> | ||
| Line 871: | Line 1,071: | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
| - | <td> | + | <td>CuMTSE2</td> |
<td>Induced</td> | <td>Induced</td> | ||
<tr> | <tr> | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
| - | <td | + | <td>pGEX6P-2</td> |
| - | <td>Non- | + | <td></td> |
| + | <td>Non-induced</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
| - | <td> | + | <td>CuMTSE1</td> |
| - | <td>Non- | + | <td>Non-induced</td> |
<tr> | <tr> | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td>Non- | + | <td>Non-induced</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--SDS-PAGE終わり--> | <!--SDS-PAGE終わり--> | ||
| Line 908: | Line 1,110: | ||
<!--Miniprep--> | <!--Miniprep--> | ||
<strong>Miniprep</strong> | <strong>Miniprep</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 920: | Line 1,123: | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
| - | <td> | + | <td>CuMTSE1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 928: | Line 1,131: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Mineprep終わり--> | <!--Mineprep終わり--> | ||
<br> | <br> | ||
<!--PCR--> | <!--PCR--> | ||
<strong>PCR</strong> | <strong>PCR</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 945: | Line 1,150: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
<td>KOD-FX-NEO</td> | <td>KOD-FX-NEO</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
| - | <td> | + | <td>CuMTSE1</td> |
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
<td>KOD-FX-NEO</td> | <td>KOD-FX-NEO</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
| - | <td> | + | <td>CuMTSE2</td> |
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
<td>KOD-FX-NEO</td> | <td>KOD-FX-NEO</td> | ||
<tr> | <tr> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
<td>KOD-FX-NEO</td> | <td>KOD-FX-NEO</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--PCR終わり--> | <!--PCR終わり--> | ||
<br> | <br> | ||
<!--AGE--> | <!--AGE--> | ||
<strong>AGE</strong> | <strong>AGE</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
<th rowspan="2">Lane</th> | <th rowspan="2">Lane</th> | ||
| - | <th colspan=" | + | <th colspan="3">Sample</th> |
<th rowspan="2">Insert</th> | <th rowspan="2">Insert</th> | ||
<th rowspan="2">Primer</th> | <th rowspan="2">Primer</th> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| + | <th>Marker</th> | ||
<th>Host</th> | <th>Host</th> | ||
<th>Vector</th> | <th>Vector</th> | ||
| Line 984: | Line 1,192: | ||
<td>1</td> | <td>1</td> | ||
<td>1kb Ladder</td> | <td>1kb Ladder</td> | ||
| - | <td | + | <td></td> |
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>2</td> | <td>2</td> | ||
| + | <td></td> | ||
<td>BY4247</td> | <td>BY4247</td> | ||
| - | <td | + | <td>p427-TEF</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>3</td> | <td>3</td> | ||
| + | <td></td> | ||
<td></td> | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
<tr> | <tr> | ||
<td>4</td> | <td>4</td> | ||
| + | <td></td> | ||
<td></td> | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
| - | <td> | + | <td>CuMTSE1</td> |
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>5</td> | <td>5</td> | ||
| + | <td></td> | ||
<td></td> | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
| - | <td> | + | <td>CuMTSE2</td> |
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>6</td> | <td>6</td> | ||
| + | <td></td> | ||
<td></td> | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--AGE終わり--> | <!--AGE終わり--> | ||
<br> | <br> | ||
| + | <img src="/wiki/images/2/29/Kit_June21_c.jpg"> | ||
<br> | <br> | ||
</p> | </p> | ||
| Line 1,037: | Line 1,257: | ||
<!--Preculture--> | <!--Preculture--> | ||
<strong>Preculture</strong> | <strong>Preculture</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,047: | Line 1,268: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>BL21(DE3)pLysS</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 1,059: | Line 1,282: | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Preculture終わり--> | <!--Preculture終わり--> | ||
<br> | <br> | ||
| Line 1,075: | Line 1,299: | ||
<!-- 実験者名終わり --> | <!-- 実験者名終わり --> | ||
<br> | <br> | ||
| - | + | Cultivate following samples in Big Scale at 30℃ overnight in shaken culture. | |
<br> | <br> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,087: | Line 1,312: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>BL21(DE3)pLysS</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 1,100: | Line 1,327: | ||
<tr> | <tr> | ||
</table> | </table> | ||
| + | </div> | ||
<br> | <br> | ||
| - | <!--DNA | + | <!--DNA Refinement 1--> |
| - | <strong>DNA | + | <strong>DNA Refinement 1</strong> |
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,115: | Line 1,344: | ||
<tr> | <tr> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td> | + | <td>CuMTSE1</td> |
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
</table> | </table> | ||
| - | <!--DNA | + | </div> |
| + | <!--DNA Refinement 1終わり--> | ||
<br> | <br> | ||
| Line 1,127: | Line 1,357: | ||
<!--Restriction Digest--> | <!--Restriction Digest--> | ||
<strong>Restriction Digest</strong> | <strong>Restriction Digest</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,141: | Line 1,372: | ||
<td></td> | <td></td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td></td> | <td></td> | ||
| - | <td> | + | <td>CuMTSE1</td> |
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
| - | <td | + | <td></td> |
| - | <td> | + | <td></td> |
| + | <td><em>Spe</em>1, <em>Xho</em>1</td> | ||
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Ristriction Digest--> | <!--Ristriction Digest--> | ||
<br> | <br> | ||
| - | <!--DNA | + | <!--DNA Refinement 2--> |
| - | <strong>DNA | + | <strong>DNA Refinement 2</strong> |
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,178: | Line 1,412: | ||
<td></td> | <td></td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td></td> | <td></td> | ||
| - | <td> | + | <td>CuMTSE1</td> |
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
| - | <td | + | <td></td> |
| - | <td> | + | <td></td> |
| + | <td><em>Spe</em>1, <em>Xho</em>1</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | <!--DNA | + | </div> |
| + | <!--DNA Refinement 2--> | ||
<br> | <br> | ||
| Line 1,199: | Line 1,435: | ||
<!--Main Culture--> | <!--Main Culture--> | ||
<strong>Main Culture</strong> | <strong>Main Culture</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,231: | Line 1,468: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td></td> | <td></td> | ||
| - | <td>Non- | + | <td>Non-induced</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 1,237: | Line 1,474: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td>Non- | + | <td>Non-induced</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 1,243: | Line 1,480: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td>Non- | + | <td>Non-induced</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Main Culture--> | <!--Main Culture--> | ||
| - | + | <img src="/wiki/images/1/1f/Kit_June25_c.jpg"> | |
<br> | <br> | ||
<!--Ligation--> | <!--Ligation--> | ||
<strong>Ligation</strong> | <strong>Ligation</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,264: | Line 1,503: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
DNA ligase: DNA Ligation Kit ‹Mighty Mix› (TAKARA BIO INC.) | DNA ligase: DNA Ligation Kit ‹Mighty Mix› (TAKARA BIO INC.) | ||
<!--Ligation--> | <!--Ligation--> | ||
| Line 1,289: | Line 1,529: | ||
<!--Transformation--> | <!--Transformation--> | ||
<strong>Transformation</strong> | <strong>Transformation</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,303: | Line 1,544: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 1,309: | Line 1,550: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 1,315: | Line 1,556: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 1,321: | Line 1,562: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Transformation--> | <!--Transformation--> | ||
<br> | <br> | ||
| Line 1,344: | Line 1,586: | ||
Isolate each colony from the plates which were inoculated on June 25. | Isolate each colony from the plates which were inoculated on June 25. | ||
<br> | <br> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,359: | Line 1,602: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
<td>1~4</td> | <td>1~4</td> | ||
</tr> | </tr> | ||
| Line 1,366: | Line 1,609: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
<td>a~h</td> | <td>a~h</td> | ||
</tr> | </tr> | ||
| Line 1,373: | Line 1,616: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
<td>ア~タ</td> | <td>ア~タ</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Colony Isolation--> | <!--Colony Isolation--> | ||
<br> | <br> | ||
| Line 1,392: | Line 1,636: | ||
<!--Colony Sweep--> | <!--Colony Sweep--> | ||
<strong>Colony Sweep</strong> | <strong>Colony Sweep</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,410: | Line 1,655: | ||
<tr> | <tr> | ||
<td>2</td> | <td>2</td> | ||
| - | <td | + | <td>1</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>3</td> | <td>3</td> | ||
| - | <td | + | <td>2</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>4</td> | <td>4</td> | ||
| - | <td | + | <td>3</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>5</td> | <td>5</td> | ||
| - | <td | + | <td>4</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>6</td> | <td>6</td> | ||
| - | <td | + | <td>a</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>7</td> | <td>7</td> | ||
| - | <td | + | <td>b</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>8</td> | <td>8</td> | ||
| - | <td | + | <td>c</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>9</td> | <td>9</td> | ||
| - | <td | + | <td>d</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>10</td> | <td>10</td> | ||
| - | <td | + | <td>e</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>11</td> | <td>11</td> | ||
| - | <td | + | <td>f</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>12</td> | <td>12</td> | ||
| - | <td | + | <td>g</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>13</td> | <td>13</td> | ||
| - | <td | + | <td>h</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Colony Sweep--> | <!--Colony Sweep--> | ||
| + | <img src="/wiki/images/4/4d/Kit_June27-1_c.jpg"> | ||
<br> | <br> | ||
<!--Colony Sweep--> | <!--Colony Sweep--> | ||
<strong>Colony Sweep</strong> | <strong>Colony Sweep</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,480: | Line 1,752: | ||
<tr> | <tr> | ||
<td>2</td> | <td>2</td> | ||
| - | <td | + | <td>ア</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>3</td> | <td>3</td> | ||
| - | <td | + | <td>イ</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>4</td> | <td>4</td> | ||
| - | <td | + | <td>ウ</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>5</td> | <td>5</td> | ||
| - | <td | + | <td>エ</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>6</td> | <td>6</td> | ||
| - | <td | + | <td>オ</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>7</td> | <td>7</td> | ||
| - | <td | + | <td>カ</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>8</td> | <td>8</td> | ||
| - | <td | + | <td>キ</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>9</td> | <td>9</td> | ||
| - | <td | + | <td>ク</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>10</td> | <td>10</td> | ||
| - | <td | + | <td>ケ</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>11</td> | <td>11</td> | ||
| - | <td | + | <td>コ</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>12</td> | <td>12</td> | ||
| - | <td | + | <td>サ</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>13</td> | <td>13</td> | ||
| - | <td | + | <td>シ</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>14</td> | <td>14</td> | ||
| - | <td | + | <td>ス</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>15</td> | <td>15</td> | ||
| - | <td | + | <td>セ</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>16</td> | <td>16</td> | ||
| - | <td | + | <td>ソ</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>17</td> | <td>17</td> | ||
| - | <td | + | <td>タ</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Colony Sweep--> | <!--Colony Sweep--> | ||
| + | <img src="/wiki/images/4/46/Kit_June27-2_c.jpg"> | ||
<br> | <br> | ||
Identified targeted genes in 4, c, h, イ, カ, セ | Identified targeted genes in 4, c, h, イ, カ, セ | ||
| Line 1,552: | Line 1,858: | ||
<!--Miniprep--> | <!--Miniprep--> | ||
<strong>Miniprep</strong> | <strong>Miniprep</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,577: | Line 1,884: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Miniprep終わり--> | <!--Miniprep終わり--> | ||
| - | |||
<br> | <br> | ||
| Line 1,584: | Line 1,891: | ||
<!--Restriction Digest--> | <!--Restriction Digest--> | ||
<strong>Restriction Digest</strong> | <strong>Restriction Digest</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,595: | Line 1,903: | ||
<tr> | <tr> | ||
<td>4</td> | <td>4</td> | ||
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>C</td> | <td>C</td> | ||
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>H</td> | <td>H</td> | ||
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
<tr> | <tr> | ||
<td>イ</td> | <td>イ</td> | ||
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>カ</td> | <td>カ</td> | ||
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Restriction Digest終わり--> | <!--Restriction Digest終わり--> | ||
| Line 1,624: | Line 1,933: | ||
<!--AGE--> | <!--AGE--> | ||
<strong>AGE</strong> | <strong>AGE</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
<th rowspan="2">Lane</th> | <th rowspan="2">Lane</th> | ||
| - | <th colspan=" | + | <th colspan="3">Sample</th> |
<th rowspan="2">Primer</th> | <th rowspan="2">Primer</th> | ||
<th rowspan="2">Enzyme</th> | <th rowspan="2">Enzyme</th> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| + | <th>Marker</th> | ||
<th>Host</th> | <th>Host</th> | ||
<th>Insert</th> | <th>Insert</th> | ||
| Line 1,637: | Line 1,948: | ||
<tr> | <tr> | ||
<td>1</td> | <td>1</td> | ||
| - | <td> | + | <td>1kb Ladder</td> |
| - | <td | + | <td></td> |
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>2</td> |
| + | <td></td> | ||
| + | <td></td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
<td></td> | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>3</td> | <td>3</td> | ||
| - | <td | + | <td></td> |
| - | <td> | + | <td>4</td> |
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Spe</em>1, <em>Xho</em>1</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>4</td> | <td>4</td> | ||
| - | <td | + | <td></td> |
| - | <td> | + | <td>4</td> |
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Spe</em>1, <em>Xho</em>1</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>5</td> | <td>5</td> | ||
| - | <td | + | <td></td> |
| - | <td> | + | <td>4</td> |
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>Non Treated</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>6</td> |
| + | <td></td> | ||
| + | <td></td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>7</td> | <td>7</td> | ||
| - | <td | + | <td></td> |
| - | <td> | + | <td>c</td> |
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Spe</em>1, <em>Xho</em>1</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>8</td> | <td>8</td> | ||
| - | <td | + | <td></td> |
| - | <td> | + | <td>c</td> |
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Spe</em>1, <em>Xho</em>1</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>9</td> | <td>9</td> | ||
| - | <td | + | <td></td> |
| - | <td> | + | <td>c</td> |
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>Non Treated</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>10</td> | <td>10</td> | ||
| - | <td | + | <td></td> |
| - | <td> | + | <td>h</td> |
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Spe</em>1, <em>Xho</em>1</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>11</td> | <td>11</td> | ||
| - | <td | + | <td></td> |
| - | <td> | + | <td>h</td> |
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Spe</em>1, <em>Xho</em>1</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>12</td> | <td>12</td> | ||
| - | <td | + | <td></td> |
| - | <td> | + | <td>h</td> |
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>Non Treated</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--AGE--> | <!--AGE--> | ||
| + | <img src="/wiki/images/9/95/June27-3_c.jpg"> | ||
<br> | <br> | ||
<!--Transformation (electroportation)--> | <!--Transformation (electroportation)--> | ||
<strong>Transformation (electroportation)</strong> | <strong>Transformation (electroportation)</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,716: | Line 2,065: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 1,722: | Line 2,071: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Transformation (electroportation)--> | <!--Transformation (electroportation)--> | ||
| Line 1,746: | Line 2,096: | ||
Divide each of 2 plates into 8 sections and inoculate following samples on them. | Divide each of 2 plates into 8 sections and inoculate following samples on them. | ||
<br> | <br> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,761: | Line 2,112: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
<td>A~H</td> | <td>A~H</td> | ||
</tr> | </tr> | ||
| Line 1,768: | Line 2,119: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
<td>一~ハ</td> | <td>一~ハ</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
Cultivate 17 samples in a small scale at 30℃ for 6 hours in shaken culture. | Cultivate 17 samples in a small scale at 30℃ for 6 hours in shaken culture. | ||
<!--Inoculation--> | <!--Inoculation--> | ||
| Line 1,781: | Line 2,133: | ||
</p> | </p> | ||
<!--本文終わり--> | <!--本文終わり--> | ||
| + | </div> | ||
| + | </div> | ||
| - | <h2>July</h2> | + | <div class="accordion"> |
| + | <h2><a href="javascript:void(0);">July</a></h2> | ||
<div class="scroll"></div> | <div class="scroll"></div> | ||
| + | <div class="ac"> | ||
| Line 1,795: | Line 2,151: | ||
KOBAYASHI<br> | KOBAYASHI<br> | ||
<!-- 実験者名終わり --> | <!-- 実験者名終わり --> | ||
| - | <!--Protein Extraction ( | + | <!--Protein Extraction (<em>S. cerevisiae</em>)--> |
| - | <strong>Protein Extraction ( | + | <strong>Protein Extraction (<em>S. cerevisiae</em>)</strong> |
| - | < | + | <div class="m_table"> |
| - | + | ||
| - | + | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,814: | Line 2,168: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 1,820: | Line 2,174: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| - | <!--Protein Extraction ( | + | </div> |
| + | <!--Protein Extraction (<em>S. cerevisiae</em>)--> | ||
<br> | <br> | ||
| Line 1,840: | Line 2,195: | ||
<strong>Miniprep</strong> | <strong>Miniprep</strong> | ||
<br> | <br> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,852: | Line 2,208: | ||
<tr> | <tr> | ||
<td>BY4247</td> | <td>BY4247</td> | ||
| - | <td | + | <td>p427-TEF</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 1,858: | Line 2,216: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 1,864: | Line 2,222: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Miniprep--> | <!--Miniprep--> | ||
| Line 1,873: | Line 2,232: | ||
<!--Restriction Digest--> | <!--Restriction Digest--> | ||
<strong>Restriction Digest</strong> | <strong>Restriction Digest</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,888: | Line 2,248: | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
<td></td> | <td></td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 1,894: | Line 2,254: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 1,901: | Line 2,261: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 1,909: | Line 2,269: | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
<td></td> | <td></td> | ||
| - | <td> | + | <td><em>Sac</em>1, <em>Kpn</em>1</td> |
<td>Buffer L</td> | <td>Buffer L</td> | ||
</tr> | </tr> | ||
| Line 1,915: | Line 2,275: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Sac</em>1, <em>Kpn</em>1</td> |
<td>Buffer L</td> | <td>Buffer L</td> | ||
</tr> | </tr> | ||
| Line 1,922: | Line 2,282: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Sac</em>1, <em>Kpn</em>1</td> |
| + | <td>Buffer L</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <!--Restriction Digest--> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <!--AGE--> | ||
| + | <strong>AGE</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Lane</th> | ||
| + | <th colspan="4">Sample</th> | ||
| + | <th rowspan="2">Enzyme</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Marker</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1</td> | ||
| + | <td></td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>Non Treated</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>2</td> | ||
| + | <td></td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTSE1</td> | ||
| + | <td>pGEX6P-2F1, pGEX6P-2R</td> | ||
| + | <td>Non Treated</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>3</td> | ||
| + | <td></td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTSE1</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | <td>Non Treated</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>4</td> | ||
| + | <td>1kb Ladder</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>5</td> | ||
| + | <td></td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>6</td> | ||
| + | <td></td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTSE1</td> | ||
| + | <td>pGEX6P-2F1, pGEX6P-2R</td> | ||
| + | <td><em>Eco</em>R1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>7</td> | ||
| + | <td></td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTSE1</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | <td><em>Eco</em>R1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>8</td> | ||
| + | <td>1kb Ladder</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>9</td> | ||
| + | <td></td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Kpn</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>10</td> | ||
| + | <td></td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTSE1</td> | ||
| + | <td>pGEX6P-2F1, pGEX6P-2R</td> | ||
| + | <td><em>Kpn</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>11</td> | ||
| + | <td></td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTSE1</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | <td><em>Kpn</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>12</td> | ||
| + | <td>1kb Ladder</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>13</td> | ||
| + | <td></td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Sac</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>14</td> | ||
| + | <td></td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTSE1</td> | ||
| + | <td>pGEX6P-2F1, pGEX6P-2R</td> | ||
| + | <td><em>Sac</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>15</td> | ||
| + | <td></td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTSE1</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | <td><em>Sac</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <!--AGE--> | ||
| + | <img src="/wiki/images/8/82/Kit_July2-1_c.jpg"> | ||
| + | <br> | ||
| + | |||
| + | <!--Restriction Digest--> | ||
| + | <strong>Restriction Digest</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="3">Sample</th> | ||
| + | <th rowspan="2">Enzyme</th> | ||
| + | <th rowspan="2">Buffer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTSE1</td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1</td> | ||
| + | <td>Buffer H</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTSE1</td> | ||
| + | <td>pGEX6P-2F1, pGEX6P-2R</td> | ||
| + | <td><em>Eco</em>R1</td> | ||
| + | <td>Buffer H</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTSE1</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | <td><em>Eco</em>R1</td> | ||
| + | <td>Buffer H</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTSE1</td> | ||
| + | <td></td> | ||
| + | <td><em>Sac</em>1</td> | ||
| + | <td>Buffer L</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTSE1</td> | ||
| + | <td>pGEX6P-2F1, pGEX6P-2R</td> | ||
| + | <td><em>Sac</em>1</td> | ||
| + | <td>Buffer L</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTSE1</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | <td><em>Sac</em>1</td> | ||
<td>Buffer L</td> | <td>Buffer L</td> | ||
</tr> | </tr> | ||
| Line 1,930: | Line 2,494: | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
<td></td> | <td></td> | ||
| - | <td> | + | <td><em>Kpn</em>1</td> |
<td>Buffer L</td> | <td>Buffer L</td> | ||
</tr> | </tr> | ||
| Line 1,936: | Line 2,500: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Kpn</em>1</td> |
<td>Buffer L</td> | <td>Buffer L</td> | ||
</tr> | </tr> | ||
| Line 1,943: | Line 2,507: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Kpn</em>1</td> |
<td>Buffer L</td> | <td>Buffer L</td> | ||
</tr> | </tr> | ||
| Line 1,951: | Line 2,515: | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
<td></td> | <td></td> | ||
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 1,957: | Line 2,521: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 1,964: | Line 2,528: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Restriction Digest--> | <!--Restriction Digest--> | ||
| Line 1,975: | Line 2,540: | ||
<!--AGE--> | <!--AGE--> | ||
<strong>AGE</strong> | <strong>AGE</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 1,982: | Line 2,548: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <th></th> | + | <th>Marker</th> |
<th>Vector</th> | <th>Vector</th> | ||
<th>Insert</th> | <th>Insert</th> | ||
| Line 1,990: | Line 2,556: | ||
<td>1</td> | <td>1</td> | ||
<td>1kb Ladder</td> | <td>1kb Ladder</td> | ||
| - | <td | + | <td></td> |
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 1,998: | Line 2,567: | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
<td></td> | <td></td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>3</td> |
| + | <td></td> | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Eco</em>R1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>4</td> |
| + | <td></td> | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Eco</em>R1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>5</td> |
| + | <td></td> | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
<td></td> | <td></td> | ||
| - | <td> | + | <td><em>Kpn</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>6</td> |
| + | <td></td> | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Kpn</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>7</td> |
| + | <td></td> | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Kpn</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>8</td> |
| + | <td></td> | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
<td></td> | <td></td> | ||
| - | <td> | + | <td><em>Sac</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>9</td> |
| + | <td></td> | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Sac</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>10</td> |
| + | <td></td> | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Sac</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>11</td> |
| + | <td></td> | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
<td></td> | <td></td> | ||
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>12</td> |
| + | <td></td> | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>13</td> |
| + | <td></td> | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--AGE--> | <!--AGE--> | ||
| - | + | <img src="/wiki/images/c/c1/Kit_July2-2_c.jpg"> | |
<br> | <br> | ||
<br> | <br> | ||
| Line 2,095: | Line 2,676: | ||
<strong>PCR</strong> | <strong>PCR</strong> | ||
<br> | <br> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 2,108: | Line 2,690: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
<td>KOD-FX-NEO</td> | <td>KOD-FX-NEO</td> | ||
</tr> | </tr> | ||
| Line 2,114: | Line 2,696: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
<td>KOD-FX-NEO</td> | <td>KOD-FX-NEO</td> | ||
</tr> | </tr> | ||
| Line 2,120: | Line 2,702: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
<td>KOD-FX-NEO</td> | <td>KOD-FX-NEO</td> | ||
</tr> | </tr> | ||
| Line 2,126: | Line 2,708: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
<td>KOD-FX-NEO</td> | <td>KOD-FX-NEO</td> | ||
</tr> | </tr> | ||
| Line 2,132: | Line 2,714: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
<td>KOD-FX-NEO</td> | <td>KOD-FX-NEO</td> | ||
</tr> | </tr> | ||
| Line 2,138: | Line 2,720: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
<td>KOD-FX-NEO</td> | <td>KOD-FX-NEO</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--PCR--> | <!--PCR--> | ||
<br> | <br> | ||
| - | <!--Protein Extraction ( | + | <!--Protein Extraction (<em>S. cerevisiae</em>)--> |
| - | <strong>Protein Extraction ( | + | <strong>Protein Extraction (<em>S. cerevisiae</em>)</strong> |
<br> | <br> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 2,166: | Line 2,750: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td> | + | <td>A</td> |
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td> | + | <td>六</td> |
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| - | <!--Protein Extraction ( | + | </div> |
| + | <!--Protein Extraction (<em>S. cerevisiae</em>)--> | ||
<br> | <br> | ||
| Line 2,194: | Line 2,779: | ||
<!--AGE--> | <!--AGE--> | ||
<strong>AGE</strong> | <strong>AGE</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
<th rowspan="2">Lane</th> | <th rowspan="2">Lane</th> | ||
| - | <th colspan="4">Sample</th> | + | <th colspan="4">Sample/Marker</th> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <th> | + | <th>Marker</th> |
<th>Vector</th> | <th>Vector</th> | ||
<th>Insert</th> | <th>Insert</th> | ||
| Line 2,208: | Line 2,794: | ||
<td>1</td> | <td>1</td> | ||
<td>1kb Ladder</td> | <td>1kb Ladder</td> | ||
| - | <td | + | <td></td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 2,215: | Line 2,803: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>3</td> |
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
| - | <td> | + | <td>CuMTSE2</td> |
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>4</td> |
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>5</td> |
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>6</td> |
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>7</td> |
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--AGE--> | <!--AGE--> | ||
| - | + | <img src="/wiki/images/9/94/Kit_July4-1_c.jpg"> | |
<br> | <br> | ||
| Line 2,255: | Line 2,849: | ||
<strong>PCR</strong> | <strong>PCR</strong> | ||
<br> | <br> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 2,268: | Line 2,863: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
<td>KOD-FX-NEO</td> | <td>KOD-FX-NEO</td> | ||
</tr> | </tr> | ||
| Line 2,274: | Line 2,869: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
<td>KOD-FX-NEO</td> | <td>KOD-FX-NEO</td> | ||
</tr> | </tr> | ||
| Line 2,280: | Line 2,875: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
<td>KOD-FX-NEO</td> | <td>KOD-FX-NEO</td> | ||
</tr> | </tr> | ||
| Line 2,286: | Line 2,881: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
<td>KOD-FX-NEO</td> | <td>KOD-FX-NEO</td> | ||
</tr> | </tr> | ||
| Line 2,292: | Line 2,887: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
<td>KOD-FX-NEO</td> | <td>KOD-FX-NEO</td> | ||
</tr> | </tr> | ||
| Line 2,298: | Line 2,893: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
<td>KOD-FX-NEO</td> | <td>KOD-FX-NEO</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--PCR--> | <!--PCR--> | ||
| Line 2,308: | Line 2,904: | ||
<!--AGE--> | <!--AGE--> | ||
<strong>AGE</strong> | <strong>AGE</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 2,314: | Line 2,911: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <th></th> | + | <th>Marker</th> |
<th>Vector</th> | <th>Vector</th> | ||
<th>Insert</th> | <th>Insert</th> | ||
| Line 2,322: | Line 2,919: | ||
<td>1</td> | <td>1</td> | ||
<td>1kb Ladder</td> | <td>1kb Ladder</td> | ||
| - | <td | + | <td></td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 2,329: | Line 2,928: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>3</td> |
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>1kb Ladder+K83:N91</td> | <td>1kb Ladder+K83:N91</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>4</td> |
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>5</td> |
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>6</td> |
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td | + | <td>7</td> |
| + | <td></td> | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--AGE--> | <!--AGE--> | ||
| - | + | <img src="/wiki/images/d/d4/Kit_July4-2_c.jpg"> | |
<br> | <br> | ||
<br> | <br> | ||
| Line 2,379: | Line 2,984: | ||
<strong>SDS-PAGE</strong> | <strong>SDS-PAGE</strong> | ||
<br> | <br> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 2,391: | Line 2,997: | ||
<tr> | <tr> | ||
<td>BL21(DE3)pLysS</td> | <td>BL21(DE3)pLysS</td> | ||
| - | <td | + | <td>pGEX6P-2</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 2,397: | Line 3,005: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 2,403: | Line 3,011: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
<tr> | <tr> | ||
<td>BY4247</td> | <td>BY4247</td> | ||
| - | <td | + | <td>p427-TEF</td> |
| + | <td></td> | ||
| + | <td></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td> | + | <td>A</td> |
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE1</td> | <td>CuMTSE1</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td> | + | <td>六</td> |
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
| - | <td> | + | <td>CuMTSE1</td> |
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--SDS-PAGE--> | <!--SDS-PAGE--> | ||
| Line 2,436: | Line 3,047: | ||
SOGA<br> | SOGA<br> | ||
<!-- 実験者名終わり --> | <!-- 実験者名終わり --> | ||
| - | <!--DNA | + | <!--DNA Refinement 1--> |
| - | <strong>DNA | + | <strong>DNA Refinement 1</strong> |
<br> | <br> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 2,448: | Line 3,060: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td> | + | <td>CuMTSE2</td> |
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
<tr> | <tr> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| - | <!--DNA | + | </div> |
| + | <!--DNA Refinement 1--> | ||
<br> | <br> | ||
| Line 2,469: | Line 3,082: | ||
<!--Restriction Digest--> | <!--Restriction Digest--> | ||
<strong>Restriction Digest</strong> | <strong>Restriction Digest</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 2,483: | Line 3,097: | ||
<td></td> | <td></td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Fast digest buffer</td> | <td>Fast digest buffer</td> | ||
</tr> | </tr> | ||
| Line 2,490: | Line 3,104: | ||
<td></td> | <td></td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Fast digest buffer</td> | <td>Fast digest buffer</td> | ||
</tr> | </tr> | ||
| Line 2,497: | Line 3,111: | ||
<td></td> | <td></td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Fast digest buffer</td> | <td>Fast digest buffer</td> | ||
</tr> | </tr> | ||
<td></td> | <td></td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Fast digest buffer</td> | <td>Fast digest buffer</td> | ||
</tr> | </tr> | ||
| Line 2,510: | Line 3,124: | ||
<td></td> | <td></td> | ||
<td></td> | <td></td> | ||
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Fast digest buffer</td> | <td>Fast digest buffer</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Ristriction Digest--> | <!--Ristriction Digest--> | ||
<br> | <br> | ||
| - | <!--DNA | + | <!--DNA Refinement 2--> |
| - | <strong>DNA | + | <strong>DNA Refinement 2</strong> |
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| - | |||
<th colspan="2">Sample</th> | <th colspan="2">Sample</th> | ||
<th rowspan="2">Enzyme</th> | <th rowspan="2">Enzyme</th> | ||
| Line 2,531: | Line 3,146: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | |||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | |||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| - | <!--DNA | + | </div> |
| + | <!--DNA Refinement 2--> | ||
<br> | <br> | ||
| Line 2,549: | Line 3,163: | ||
<!--Ligation--> | <!--Ligation--> | ||
<strong>Ligation</strong> | <strong>Ligation</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 2,562: | Line 3,177: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<br> | <br> | ||
DNA ligase: DNA Ligation Kit ‹Mighty Mix› (TAKARA BIO INC.) | DNA ligase: DNA Ligation Kit ‹Mighty Mix› (TAKARA BIO INC.) | ||
<br> | <br> | ||
| - | <!--DNA | + | <!--DNA Refinement 2--> |
<br> | <br> | ||
| - | <!--Transformation (E.coli)--> | + | <!--Transformation (<em>E. coli</em>)--> |
| - | <strong>Transformation (E.coli)</strong> | + | <strong>Transformation (<em>E. coli</em>)</strong> |
<br> | <br> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 2,597: | Line 3,214: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 2,604: | Line 3,221: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 2,611: | Line 3,228: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 2,618: | Line 3,235: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| - | <!--Transformation (E.coli)--> | + | </div> |
| + | <!--Transformation (<em>E. coli</em>)--> | ||
</p> | </p> | ||
<!--本文終わり--> | <!--本文終わり--> | ||
| Line 2,643: | Line 3,261: | ||
Isolate each colony from the plates which were inoculated on July 6. | Isolate each colony from the plates which were inoculated on July 6. | ||
<br> | <br> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 2,658: | Line 3,277: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
<td>1~16</td> | <td>1~16</td> | ||
</tr> | </tr> | ||
| Line 2,665: | Line 3,284: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
<td>ア~エ</td> | <td>ア~エ</td> | ||
</tr> | </tr> | ||
| Line 2,672: | Line 3,291: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
<td>a~p</td> | <td>a~p</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Colony Isolation--> | <!--Colony Isolation--> | ||
<br> | <br> | ||
| - | <!--Transformation (E.coli)--> | + | <!--Transformation (<em>E. coli</em>)--> |
| - | <strong>Transformation (E.coli)</strong> | + | <strong>Transformation (<em>E. coli</em>)</strong> |
<br> | <br> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 2,698: | Line 3,319: | ||
<td>p427-TEF</td> | <td>p427-TEF</td> | ||
<td>CuMTSE2</td> | <td>CuMTSE2</td> | ||
| - | <td> | + | <td>pGEX6P-2F2, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| - | <!--Transformation (E.coli)--> | + | </div> |
| + | <!--Transformation (<em>E. coli</em>)--> | ||
<br> | <br> | ||
| Line 2,718: | Line 3,340: | ||
<!--Colony Sweep--> | <!--Colony Sweep--> | ||
<strong>Colony Sweep</strong> | <strong>Colony Sweep</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 2,813: | Line 3,436: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Colony Sweep--> | <!--Colony Sweep--> | ||
| - | + | <img src="/wiki/images/e/e6/Kit_July8-1_c.jpg"> | |
<br> | <br> | ||
<!--Colony Sweep--> | <!--Colony Sweep--> | ||
<strong>Colony Sweep</strong> | <strong>Colony Sweep</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 2,854: | Line 3,479: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Colony Sweep--> | <!--Colony Sweep--> | ||
| - | + | <img src="/wiki/images/c/c1/Kit_July8-2_c.jpg"> | |
<br> | <br> | ||
<!--Colony Sweep--> | <!--Colony Sweep--> | ||
<strong>Colony Sweep</strong> | <strong>Colony Sweep</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 2,955: | Line 3,582: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Colony Sweep--> | <!--Colony Sweep--> | ||
| - | + | <img src="/wiki/images/c/c3/Kit_July8-3_c.jpg"> | |
<br> | <br> | ||
<br> | <br> | ||
| Line 2,971: | Line 3,599: | ||
<!--Main Culture--> | <!--Main Culture--> | ||
<strong>Main Culture</strong> | <strong>Main Culture</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 2,979: | Line 3,608: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td> | + | <td>d</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td> | + | <td>h</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td> | + | <td>l</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 2,997: | Line 3,626: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Main Culture--> | <!--Main Culture--> | ||
| Line 3,003: | Line 3,633: | ||
<!--Mini Prep--> | <!--Mini Prep--> | ||
<strong>Mini Prep</strong> | <strong>Mini Prep</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 3,029: | Line 3,660: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Mini Prep--> | <!--Mini Prep--> | ||
| Line 3,035: | Line 3,667: | ||
<!--Restriction Digest--> | <!--Restriction Digest--> | ||
<strong>Restriction Digest</strong> | <strong>Restriction Digest</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 3,049: | Line 3,682: | ||
<td>p427-TEF(d)</td> | <td>p427-TEF(d)</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Fast digest buffer</td> | <td>Fast digest buffer</td> | ||
</tr> | </tr> | ||
| Line 3,057: | Line 3,690: | ||
<td>p427-TEF(h)</td> | <td>p427-TEF(h)</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Fast digest buffer</td> | <td>Fast digest buffer</td> | ||
</tr> | </tr> | ||
| Line 3,065: | Line 3,698: | ||
<td>p427-TEF(l)</td> | <td>p427-TEF(l)</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Fast digest buffer</td> | <td>Fast digest buffer</td> | ||
</tr> | </tr> | ||
| Line 3,072: | Line 3,705: | ||
<td>p427-TEF(ア)</td> | <td>p427-TEF(ア)</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Fast digest buffer</td> | <td>Fast digest buffer</td> | ||
</tr> | </tr> | ||
| Line 3,080: | Line 3,713: | ||
<td>p427-TEF(イ)</td> | <td>p427-TEF(イ)</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Fast digest buffer</td> | <td>Fast digest buffer</td> | ||
</tr> | </tr> | ||
| Line 3,088: | Line 3,721: | ||
<td>p427-TEF(エ)</td> | <td>p427-TEF(エ)</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
<td>Fast digest buffer</td> | <td>Fast digest buffer</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<!--Restriction Digest--> | <!--Restriction Digest--> | ||
| Line 3,099: | Line 3,733: | ||
<!--AGE--> | <!--AGE--> | ||
<strong>AGE</strong> | <strong>AGE</strong> | ||
| + | <div class="m_table"> | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| Line 3,112: | Line 3,747: | ||
<tr> | <tr> | ||
<td>1</td> | <td>1</td> | ||
| - | <td> | + | <td>p427-TEF(ア)</td> |
| - | <td | + | <td>CuMTS3</td> |
| + | <td>pGEX6P-2F1, pGEX6P-2R</td> | ||
| + | <td>Non Treated</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 3,119: | Line 3,756: | ||
<td>p427-TEF(ア)</td> | <td>p427-TEF(ア)</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>3</td> | <td>3</td> | ||
| - | <td>p427-TEF( | + | <td>p427-TEF(イ)</td> |
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td>Non Treated</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 3,133: | Line 3,770: | ||
<td>p427-TEF(イ)</td> | <td>p427-TEF(イ)</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>5</td> | <td>5</td> | ||
| - | <td>p427-TEF( | + | <td>p427-TEF(エ)</td> |
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td>Non Treated</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 3,147: | Line 3,784: | ||
<td>p427-TEF(エ)</td> | <td>p427-TEF(エ)</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>7</td> | <td>7</td> | ||
| - | <td>p427-TEF( | + | <td>p427-TEF(d)</td> |
| - | <td> | + | <td>CuMTS62</td> |
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td>Non Treated</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 3,161: | Line 3,798: | ||
<td>p427-TEF(d)</td> | <td>p427-TEF(d)</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>9</td> | <td>9</td> | ||
| - | <td>p427-TEF( | + | <td>p427-TEF(h)</td> |
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td>Non Treated</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 3,175: | Line 3,812: | ||
<td>p427-TEF(h)</td> | <td>p427-TEF(h)</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>11</td> | <td>11</td> | ||
| - | <td>p427-TEF( | + | <td>p427-TEF(l)</td> |
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td>Non Treated</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 3,189: | Line 3,826: | ||
<td>p427-TEF(l)</td> | <td>p427-TEF(l)</td> | ||
<td>CuMTS62</td> | <td>CuMTS62</td> | ||
| - | <td> | + | <td>pGEX6P-2F1, pGEX6P-2R</td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xho</em>1</td> |
| - | </ | + | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
Marker: 1kb Ladder | Marker: 1kb Ladder | ||
<br> | <br> | ||
| + | <img src="/wiki/images/5/59/Kit_July9_c.jpg"> | ||
<!--AGE--> | <!--AGE--> | ||
| + | <br> | ||
| + | </p> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="accordion"> | ||
| + | <h2><a href="javascript:void(0);">August</a></h2> | ||
| + | <div class="scroll"></div> | ||
| + | <div class="ac"> | ||
| + | <h3>August 19, 2014</h3> | ||
| + | <p class="sentence"> | ||
| + | KOBAYASHI | ||
| + | <br> | ||
| + | Conduct mutagenesis to remove an illegal restriction site (<em>Eco</em>R1) | ||
| + | <br> | ||
| + | <strong>InversePCR</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="4">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | <th>DNA polymerase</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>CuMTS3</td> | ||
| + | <td>Terpinene3-mutagenesis-FP, Terpinene3-mutagenesis-RP</td> | ||
| + | <td>KOD-Plus</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | (TOYOBO, KOD-Plus-Mutagenesis Kit) | ||
| + | Start from KOD -Plus- Mutagenesis Kit protocol No.2. | ||
| + | <br> | ||
| + | <br> | ||
| + | </p> | ||
| - | <h3> | + | |
| + | <h3>August 20, 2014</h3> | ||
| + | <p class="sentence"> | ||
| + | KOBAYASHI | ||
<br> | <br> | ||
| - | + | Restart mutagenesis from KOD -Plus- Mutagenesis Kit protocol No.3 to obtain plasmid (muta-CuMTS3 in pGEX-6P-2) | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | < | + | <strong>Transformation</strong> |
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="4">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td rowspan="2">DH5α</td> | ||
| + | <td rowspan="2">pGEX6P-2</td> | ||
| + | <td rowspan="2">muta-CuMTS3</td> | ||
| + | <td>Terpinene3-mutagenesis-FP and Terpinene3-mutagenesis-RP</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br> | <br> | ||
| - | </p> | + | </p> |
| - | < | + | |
| - | + | <h3>August 21, 2014</h3> | |
<p class="sentence"> | <p class="sentence"> | ||
| + | KOBAYASHI | ||
<br> | <br> | ||
| + | <strong>Colony Isolation</strong><br> | ||
| + | Isolate each colony from two plates which were inoculated overnight | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="4">Sample</th> | ||
| + | <th rowspan="2">Section</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>Terpinene3-mutagenesis-FP,<br>Terpinene3-mutagenesis-RP</td> | ||
| + | <td>あ-み</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | Select 5 isolated colonies whose code names are あ,か,さ,た and な and miniprep them. | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br> | <br> | ||
| - | < | + | </p> |
| - | < | + | |
| - | < | + | <h3>August 22, 2014</h3> |
| - | < | + | <p class="sentence"> |
| - | + | KOBAYASHI | |
<br> | <br> | ||
| - | < | + | <strong>Restriction Digest</strong> |
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="3">Sample</th> | ||
| + | <th rowspan="2">Enzyme</th> | ||
| + | <th rowspan="2">Buffer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α あ</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Sal</em>1</td> | ||
| + | <td>Fast Digest buffer</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α か</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Sal</em>1</td> | ||
| + | <td>Fast Digest buffer</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α さ</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Sal</em>1</td> | ||
| + | <td>Fast Digest buffer</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α た</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Sal</em>1</td> | ||
| + | <td>Fast Digest buffer</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </table> | ||
| + | </div> | ||
<br> | <br> | ||
| - | < | + | <strong>AGE</strong> |
| - | < | + | <div class="m_table"> |
| - | < | + | <table class="materials"> |
| - | < | + | <tr> |
| - | < | + | <th rowspan="2">Lane</th> |
| + | <th colspan="4">Sample</th> | ||
| + | <th rowspan="2">Enzyme</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Marker</th> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1</td> | ||
| + | <td>1kb Ladder</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>2</td> | ||
| + | <td></td> | ||
| + | <td>DH5α あ</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Sal</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>3</td> | ||
| + | <td></td> | ||
| + | <td>DH5α か</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Sal</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>4</td> | ||
| + | <td></td> | ||
| + | <td>DH5α さ</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Sal</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>5</td> | ||
| + | <td></td> | ||
| + | <td>DH5α た</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Sal</em>1</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
<br> | <br> | ||
| - | < | + | <strong>Restriction Digest</strong> |
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="3">Sample</th> | ||
| + | <th rowspan="2">Enzyme</th> | ||
| + | <th rowspan="2">Buffer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α か</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>CuMTS3</td> | ||
| + | <td><em>Eco</em>R1</td> | ||
| + | <td>Fast Digest buffer</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α か</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Eco</em>R1</td> | ||
| + | <td>Fast Digest buffer</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
<br> | <br> | ||
| - | < | + | <strong>AGE</strong> |
| - | < | + | <div class="m_table"> |
| - | < | + | <table class="materials"> |
| - | < | + | <tr> |
| - | <br> | + | <th rowspan="2">Lane</th> |
| + | <th colspan="4">Sample</th> | ||
| + | <th rowspan="2">Enzyme</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Marker</th> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1</td> | ||
| + | <td>1kb Ladder</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>2</td> | ||
| + | <td></td> | ||
| + | <td>DH5α か</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>CuMTS3</td> | ||
| + | <td><em>Eco</em>R1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>3</td> | ||
| + | <td></td> | ||
| + | <td>DH5α か</td> | ||
| + | <td>pGEX6P-2</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Eco</em>R1</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <img src="/wiki/images/b/bc/Kit_August22_c.jpg"> | ||
| + | An illegal <em>Eco</em>R1 restriction site was removed.<br> | ||
<br> | <br> | ||
| - | < | + | <strong>PCR</strong> |
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="3">Sample</th> | ||
| + | <th rowspan="2">DNA polymerase</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>か pGEX6P-2</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>terpinene3-prefix and terpinene3-suffix</td> | ||
| + | <td>KOD-FX-NEO-Plus</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
<br> | <br> | ||
| - | < | + | <strong>AGE</strong> |
| - | < | + | <div class="m_table"> |
| - | < | + | <table class="materials"> |
| - | < | + | <tr> |
| - | < | + | <th rowspan="2">Lane</th> |
| + | <th colspan="4">Sample</th> | ||
| + | <th rowspan="2">DNA polymerase</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Marker</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1</td> | ||
| + | <td>1kb Ladder</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>2</td> | ||
| + | <td></td> | ||
| + | <td>か pGEX6P-2</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>terpinene3-prefix and terpinene3-suffix</td> | ||
| + | <td>KOD-FX-NEO-Plus</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <img src="/wiki/images/7/72/Kit_August22_1_c.jpg"> | ||
<br> | <br> | ||
<br> | <br> | ||
| + | </p> | ||
| + | |||
| + | <h3>August 30, 2014</h3> | ||
| + | KOBAYASHI | ||
<br> | <br> | ||
| - | <br></p> | + | <strong>PCR</strong> |
| - | <h2>September</h2> | + | <div class="m_table"> |
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="3">Sample</th> | ||
| + | <th rowspan="2">DNA polymerase</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>か pGEX6P-2</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>terpinene3-prefix-<em>Xba</em>1 and terpinene3-suffix-<em>Spe</em>1</td> | ||
| + | <td><em>Taq</em></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | <strong>AGE</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Lane</th> | ||
| + | <th colspan="4">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Marker</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1</td> | ||
| + | <td>1kb Ladder</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>2</td> | ||
| + | <td></td> | ||
| + | <td>か pGEX6P-2</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>terpinene3-prefix-<em>Xba</em>1 and terpinene3-suffix-<em>Spe</em>1</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | <img src="/wiki/images/3/31/Kit_August30_c.jpg"> | ||
| + | Made total amount of 295 μL of PCR samples. | ||
| + | <br> | ||
| + | </tr> | ||
| + | <br> | ||
| + | </p> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div class="accordion"> | ||
| + | <h2><a href="javascript:void(0);">September</a></h2> | ||
<div class="scroll"></div> | <div class="scroll"></div> | ||
| + | <div class="ac"> | ||
<p class="sentence"> | <p class="sentence"> | ||
| + | <h3>September 18, 2014</h3> | ||
| + | KOBAYASHI | ||
<br> | <br> | ||
| + | <strong>Miniprep</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="3">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
<br> | <br> | ||
| - | < | + | <strong>Restriction Digest</strong> |
| - | < | + | <div class="m_table"> |
| - | < | + | <table class="materials"> |
| - | < | + | <tr> |
| - | < | + | <th colspan="3">Sample</th> |
| - | < | + | <th rowspan="2">Buffer</th> |
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Enzyme</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | <td>Buffer H</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td><em>Xba</em>1, <em>Spe</em>1</td> | ||
| + | <td>Fast Digest buffer</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | </tr> | ||
<br> | <br> | ||
| - | < | + | <strong>AGE</strong> |
| - | < | + | <div class="m_table"> |
| - | < | + | <table class="materials"> |
| - | < | + | <tr> |
| - | < | + | <th rowspan="2">Lane</th> |
| + | <th colspan="4">Sample</th> | ||
| + | <th rowspan="2">Enzyme</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Marker</th> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1</td> | ||
| + | <td>1kb Ladder</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>2</td> | ||
| + | <td></td> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>3</td> | ||
| + | <td></td> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td><em>Xba</em>1, <em>Spe</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>4</td> | ||
| + | <td></td> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <img src="/wiki/images/2/29/Kit_September18_c.jpg"> | ||
<br> | <br> | ||
| - | < | + | <strong>PCR</strong> |
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="3">Sample</th> | ||
| + | <th rowspan="2">DNA polymerase</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>か pGEX6P-2</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>terpinene3-prefix-<em>Xba</em>1 and terpinene3-suffix-<em>Spe</em>1</td> | ||
| + | <td><em>Taq</em></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
<br> | <br> | ||
| - | < | + | <strong>DNA Refinement 1</strong> |
| - | < | + | <div class="m_table"> |
| - | < | + | <table class="materials"> |
| - | < | + | <tr> |
| - | < | + | <th colspan="2">Sample</th> |
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>terpinene3-prefix-<em>Xba</em>1 and terpinene3-suffix-<em>Spe</em>1</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | </tr> | ||
<br> | <br> | ||
| - | < | + | <strong>Restriction Digest</strong> |
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="3">Sample</th> | ||
| + | <th rowspan="2">Enzyme</th> | ||
| + | <th rowspan="2">Buffer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB1C3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Xba</em>1, <em>Spe</em>1</td> | ||
| + | <td>Fast Digest</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>か pGEX6P-2</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>terpinene3-prefix-<em>Xba</em>1 and terpinene3-suffix-<em>Spe</em>1</td> | ||
| + | <td><em>Xba</em>1, <em>Spe</em>1</td> | ||
| + | <td>Fast Digest</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | </tr> | ||
<br> | <br> | ||
| - | < | + | <strong>DNA Refinement 2</strong> |
| - | < | + | <div class="m_table"> |
| - | < | + | <table class="materials"> |
| - | < | + | <tr> |
| - | < | + | <th colspan="2">Sample</th> |
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>terpinene3-prefix-<em>Xba</em>1 and terpinene3-suffix-<em>Spe</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | </tr> | ||
<br> | <br> | ||
| - | < | + | <strong>Ligation</strong> |
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="3">Sample</th> | ||
| + | <th rowspan="2">Enzyme</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>DNA</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>terpinene3-prefix-<em>Xba</em>1 and terpinene3-suffix-<em>Spe<em>1</td> | ||
| + | <td><em>Spe</em>1, <em>Xba</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | </tr> | ||
| + | DNA ligase: DNA Ligation Kit ‹Mighty Mix› (TAKARA BIO INC.) | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br> | <br> | ||
| + | <strong>Transformation</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="4">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>terpinene3-prefix-<em>Xba</em>1 and terpinene3-suffix-<em>Spe</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | </tr> | ||
<br> | <br> | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <h3>September 19, 2014</h3> | ||
| + | KOBAYASHI | ||
| + | <br> | ||
| + | <strong>Colony Isolation</strong> | ||
| + | <br> | ||
| + | Isolate each colony from the plates which were inoculated overnight. | ||
| + | <br> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="4">Sample</th> | ||
| + | <th rowspan="2">Section</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>terpinene3-prefix-<em>Xba</em>1 and terpinene3-suffix-<em>Spe</em>1</td> | ||
| + | <td>A-P</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>terpinene3-prefix-<em>Xba</em>1 and terpinene3-suffix-<em>Spe</em>1</td> | ||
| + | <td>ア-タ</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | <strong>Colony Sweep</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Lane</th> | ||
| + | <th colspan="3">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1</td> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>2</td> | ||
| + | <td>ア</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>3</td> | ||
| + | <td>イ</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>4</td> | ||
| + | <td>ウ</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>5</td> | ||
| + | <td>エ</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>6</td> | ||
| + | <td>オ</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>7</td> | ||
| + | <td>カ</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>8</td> | ||
| + | <td>キ</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>9</td> | ||
| + | <td>ク</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>10</td> | ||
| + | <td>ケ</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>11</td> | ||
| + | <td>コ</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>12</td> | ||
| + | <td>サ</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>13</td> | ||
| + | <td>シ</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>14</td> | ||
| + | <td>ス</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>15</td> | ||
| + | <td>セ</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>16</td> | ||
| + | <td>ソ</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>17</td> | ||
| + | <td>タ</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <img src="/wiki/images/4/40/Kit_September19_c.jpg"> | ||
| + | <br> | ||
| + | Targeted colonies were identified in セ and ソ. | ||
| + | <br> | ||
| + | <br> | ||
| + | <strong>Colony Sweep</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Lane</th> | ||
| + | <th colspan="3">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1</td> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>2</td> | ||
| + | <td>A</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>3</td> | ||
| + | <td>B</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>4</td> | ||
| + | <td>C</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>5</td> | ||
| + | <td>D</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>6</td> | ||
| + | <td>E</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>7</td> | ||
| + | <td>F</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>8</td> | ||
| + | <td>G</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>9</td> | ||
| + | <td>H</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>10</td> | ||
| + | <td>I</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>11</td> | ||
| + | <td>J</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>12</td> | ||
| + | <td>K</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>13</td> | ||
| + | <td>L</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>14</td> | ||
| + | <td>M</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>15</td> | ||
| + | <td>N</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>16</td> | ||
| + | <td>O</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>17</td> | ||
| + | <td>P</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | </tr> | ||
| + | |||
| + | <img src="/wiki/images/e/e6/Kit_September19_1_c.jpg"> | ||
| + | <br> | ||
| + | Targeted colonies were identified in colony N and F. | ||
| + | <br> | ||
| + | <br> | ||
| + | Select セ, ソ, N, F | ||
| + | <br> | ||
| + | <br> | ||
| + | <strong>Main Culture</strong> | ||
| + | <br> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="4">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α(N)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>----------</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α(F)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>----------</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α(セ)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>----------</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α(ソ)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>----------</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | Cultivate these samples in 3mL of LB medium with ChlPhe at 30°C overnight. | ||
| + | <br> | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <h3>September 20, 2014</h3> | ||
| + | KOBAYASHI | ||
| + | <br> | ||
| + | <strong>Miniprep</strong> | ||
| + | <br> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="3">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α(N)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α(F)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α(セ)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α(ソ)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | Enzyme: E/P and <em>Xba</em>1 | ||
| + | <br> | ||
| + | <br> | ||
| + | <strong>Restriction Digest</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="4">Sample</th> | ||
| + | <th rowspan="2">Enzyme</th> | ||
| + | <th rowspan="2">Buffer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Marker</th> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>DH5α (セ)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>DH5α (ソ)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>DH5α (F)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>DH5α (N)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>DH5α (セ)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | <td>Buffer H</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>DH5α (ソ)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | <td>Buffer H</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>DH5α (F)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | <td>Buffer H</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>DH5α (N)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | <td>Buffer H</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | <td>Buffer H</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td></td> | ||
| + | <td><em>Xba</em>1</td> | ||
| + | <td>FD buffer</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>DH5α (セ)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Xba</em>1</td> | ||
| + | <td>FD buffer</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>DH5α (ソ)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Xba</em>1</td> | ||
| + | <td>FD buffer</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>DH5α (F)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Xba</em>1</td> | ||
| + | <td>FD buffer</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>DH5α (N)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Xba</em>1</td> | ||
| + | <td>FD buffer</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | <strong>AGE</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Lane</th> | ||
| + | <th colspan="4">Sample</th> | ||
| + | <th rowspan="2">Enzyme</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Marker</th> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>2</td> | ||
| + | <td></td> | ||
| + | <td>DH5α (セ)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>3</td> | ||
| + | <td></td> | ||
| + | <td>DH5α (ソ)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>4</td> | ||
| + | <td></td> | ||
| + | <td>DH5α (F)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>5</td> | ||
| + | <td></td> | ||
| + | <td>DH5α (N)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>6</td> | ||
| + | <td>1kb Ladder</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>7</td> | ||
| + | <td></td> | ||
| + | <td>DH5α (セ)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>8</td> | ||
| + | <td></td> | ||
| + | <td>DH5α (ソ)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>9</td> | ||
| + | <td></td> | ||
| + | <td>DH5α (F)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>10</td> | ||
| + | <td></td> | ||
| + | <td>DH5α (N)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>11</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>12</td> | ||
| + | <td>1kb Ladder</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>13</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td></td> | ||
| + | <td><em>Xba</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>14</td> | ||
| + | <td></td> | ||
| + | <td>DH5α (セ)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Xba</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>15</td> | ||
| + | <td></td> | ||
| + | <td>DH5α (ソ)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Xba</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>16</td> | ||
| + | <td></td> | ||
| + | <td>DH5α (F)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Xba</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>17</td> | ||
| + | <td></td> | ||
| + | <td>DH5α (N)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td><em>Xba</em>1</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | <img src="/wiki/images/e/e5/Kit_September20_c.jpg"> | ||
| + | <div class="clear"><hr /></div> | ||
| + | Sample F was selected as a submission plasmid. | ||
| + | <br> | ||
| + | <br> | ||
| + | <strong>Main Culture</strong> | ||
| + | <br> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="4">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α(F)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>----------</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α(F)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>----------</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α(F)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td>----------</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | Cultivate these samples in 3mL of LB medium with ChlPhe at 30°C overnight. | ||
| + | <br> | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <h3>September 21, 2014</h3> | ||
| + | KOBAYASHI | ||
| + | <br> | ||
| + | <strong>Miniprep</strong> | ||
| + | <br> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="4">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α(F)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α(F)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α(F)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | <strong>Restriction Digest</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="5">Sample</th> | ||
| + | <th rowspan="2">Enzyme</th> | ||
| + | <th rowspan="2">Buffer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Marker</th> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1kb Ladder</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1</td> | ||
| + | <td>Buffer H</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td></td> | ||
| + | <td><em>Pst</em>1</td> | ||
| + | <td>Buffer H</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | <td>Buffer H</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td></td> | ||
| + | <td><em>Spe</em>1</td> | ||
| + | <td>FD buffer</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td></td> | ||
| + | <td><em>Xba</em>1</td> | ||
| + | <td>FD buffer</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td></td> | ||
| + | <td><em>Spe</em>1, <em>Xba</em>1</td> | ||
| + | <td>FD buffer</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>λDNA-HindⅢ</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>F</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1</td> | ||
| + | <td>Buffer H</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>F</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | <td><em>Pst</em>1</td> | ||
| + | <td>Buffer H</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>F</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | <td>Buffer H</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>F</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | <td><em>Spe</em>1</td> | ||
| + | <td>FD buffer</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>F</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | <td><em>Xba</em>1</td> | ||
| + | <td>FD buffer</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>F</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | <td><em>Spe</em>1, <em>Xba</em>1</td> | ||
| + | <td>FD buffer</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>F</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | <strong>AGE</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Lane</th> | ||
| + | <th colspan="5">Sample</th> | ||
| + | <th rowspan="2">Enzyme</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Marker</th> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1</td> | ||
| + | <td>1kb Ladder</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>2</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td></td> | ||
| + | <td><em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>4</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>5</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td></td> | ||
| + | <td><em>Spe</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>6</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td></td> | ||
| + | <td><em>Xba</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>7</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td></td> | ||
| + | <td><em>Spe</em>1, <em>Xba</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>8</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>BBa_J04450</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>9</td> | ||
| + | <td>λDNA-HindⅢ</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>10</td> | ||
| + | <td></td> | ||
| + | <td>F</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>11</td> | ||
| + | <td></td> | ||
| + | <td>F</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | <td><em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>12</td> | ||
| + | <td></td> | ||
| + | <td>F</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>13</td> | ||
| + | <td></td> | ||
| + | <td>F</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | <td><em>Spe</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>14</td> | ||
| + | <td></td> | ||
| + | <td>F</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | <td><em>Xba</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>15</td> | ||
| + | <td></td> | ||
| + | <td>F</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | <td><em>Spe</em>1, <em>Xba</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>16</td> | ||
| + | <td></td> | ||
| + | <td>F</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>muta-CuMTS3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <img src="/wiki/images/d/d7/Kit_September21_c.jpg"> | ||
| + | |||
<br> | <br> | ||
<br></p> | <br></p> | ||
| - | <h2>October</h2> | + | </div> |
| + | </div> | ||
| + | |||
| + | <div class="accordion"> | ||
| + | <h2><a href="javascript:void(0);">October</a></h2> | ||
<div class="scroll"></div> | <div class="scroll"></div> | ||
| + | <div class="ac"> | ||
<p class="sentence"> | <p class="sentence"> | ||
| + | |||
| + | <h3>October 1, 2014</h3> | ||
| + | KOBAYASHI | ||
<br> | <br> | ||
| + | <strong>Protein Extraction (<em>S. cerevisiae</em>)</strong> | ||
<br> | <br> | ||
| - | < | + | <div class="m_table"> |
| - | < | + | <table class="materials"> |
| - | < | + | <tr> |
| - | < | + | <th colspan="4">Sample</th> |
| - | < | + | </tr> |
| - | < | + | <tr> |
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTSE2</td> | ||
| + | <td>pGEX6P-2F1, pGEX6P-2R</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTS3</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTS62</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br> | <br> | ||
| - | < | + | <strong>SDS-PAGE</strong> |
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Lane</th> | ||
| + | <th colspan="4">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1</td> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>2</td> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTSE2</td> | ||
| + | <td>pGEX6P-2F1, pGEX6P-2R</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>3</td> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTS3</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>4</td> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTS62</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
<br> | <br> | ||
| - | < | + | </tr> |
| - | < | + | </p> |
| - | < | + | |
| - | < | + | |
| - | + | <h3>October 2, 2014</h3> | |
| + | KOBAYASHI | ||
<br> | <br> | ||
| - | < | + | <strong>Preculture</strong> |
<br> | <br> | ||
| - | < | + | <div class="m_table"> |
| - | < | + | <table class="materials"> |
| - | < | + | <tr> |
| - | < | + | <th colspan="4">Sample</th> |
| - | < | + | </tr> |
| + | <tr> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTS3</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
<br> | <br> | ||
| - | + | Cultivate these samples in 3mL of SD medium with G418 at 30°C overnight. | |
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br> | <br> | ||
| + | <strong>Western blot</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Lane</th> | ||
| + | <th colspan="4">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1</td> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>2</td> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTSE2</td> | ||
| + | <td>pGEX6P-2F1, pGEX6P-2R</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>3</td> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTS3</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>4</td> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTS62</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
<br> | <br> | ||
| + | <img src="/wiki/images/e/e0/Kit_October2_c.jpg"> | ||
| + | </p> | ||
| + | |||
| + | <h3>October 3, 2014</h3> | ||
| + | KOBAYASHI | ||
<br> | <br> | ||
| - | <br></p> | + | <strong>Main culture</strong> |
| + | <br> | ||
| + | Cultivate following samples in Big Scale with G418 at 28°C overnight in shaken culture. | ||
| + | <br> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="4">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTS3</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | <strong>Restriction Digest</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="3">Sample</th> | ||
| + | <th rowspan="2">Enzyme</th> | ||
| + | <th rowspan="2">Buffer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB1C3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1, <em>Spe</em>1</td> | ||
| + | <td>Buffer H</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB1C3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Xba</em>1, <em>Pst</em>1</td> | ||
| + | <td>Buffer H</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Linearized pSB1A3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | <td>Buffer H</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | <strong>AGE</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Lane</th> | ||
| + | <th rowspan="2">Marker</th> | ||
| + | <th colspan="3">Sample</th> | ||
| + | <th rowspan="2">Enzyme</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1</td> | ||
| + | <td>1kb Ladder</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>2</td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1, <em>Spe</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>3</td> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Xba</em>1, <em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>4</td> | ||
| + | <td></td> | ||
| + | <td>Linearized pSB1A3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | <strong>DNA Refinement 1</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="3">Sample</th> | ||
| + | <th rowspan="2">Enzyme</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB1C3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1, <em>Spe</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB1C3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Xba</em>1, <em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB1C3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Xba</em>1, <em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Linearized pSB1A3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1, <em>Dpn</em>1</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | <strong>Ligation</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Vector</th> | ||
| + | <th colspan="2">Sample</th> | ||
| + | <th rowspan="2">Enzyme</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Insert1</th> | ||
| + | <th>Insert2</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB1A3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1, <em>Xba</em>1, <em>Spe</em>1, <em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB1A3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1, <em>Xba</em>1, <em>Spe</em>1, <em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | DNA ligase: DNA Ligation Kit ‹Mighty Mix› (TAKARA BIO INC.) | ||
| + | <br> | ||
| + | <br> | ||
| + | <strong>Transformation</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Host</th> | ||
| + | <th colspan="3">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert1</th> | ||
| + | <th>Insert2</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1A3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1A3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | </tr> | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <h3>October 4, 2014</h3> | ||
| + | KOBAYASHI | ||
| + | <br> | ||
| + | <strong>Colony Isolation</strong> | ||
| + | <br> | ||
| + | Isolate each colony from the plates which were inoculated overnight. | ||
| + | <br> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Host</th> | ||
| + | <th colspan="3">Sample</th> | ||
| + | <th rowspan="2">Section</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1A3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>1-16</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1A3</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td>a-p</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | <strong>Monoterpene Extraction</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Host</th> | ||
| + | <th colspan="3">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTS3</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | Samples were stocked in -4℃. | ||
| + | <br> | ||
| + | <br> | ||
| + | <strong>Preculture</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <th rowspan="2">Host</th> | ||
| + | <th colspan="3">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTSE2</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTS62</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | Cultivate samples in 3mL of SD medium with G418 at 30°C overnight. | ||
| + | <br> | ||
| + | </tr> | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <h3>October 5, 2014</h3> | ||
| + | KOBAYASHI | ||
| + | <br> | ||
| + | <strong>Miniprep</strong> | ||
| + | <br> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Host</th> | ||
| + | <th colspan="2">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1A3</td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1A3</td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | <strong>Restriction Digest and AGE</strong> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Host</th> | ||
| + | <th colspan="2">Sample</th> | ||
| + | <th rowspan="2">Enzyme</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td></td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>K1448000</td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | </tr> | ||
| + | </p> | ||
| + | |||
| + | <h3>October 8, 2014</h3> | ||
| + | KOBAYASHI | ||
| + | <br> | ||
| + | <strong>GC-MS analysis</strong> | ||
| + | <br> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Host</th> | ||
| + | <th colspan="3">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTS3</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | <strong>Preculture</strong> | ||
| + | <br> | ||
| + | <div class="m_table"> | ||
| + | |||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Host</th> | ||
| + | <th colspan="3">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTS3</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | </tr> | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <h3>October 9, 2014</h3> | ||
| + | KOBAYASHI | ||
| + | <br> | ||
| + | <strong>Main Culture</strong> | ||
| + | <br> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Host</th> | ||
| + | <th colspan="3">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTS3</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | </tr> | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <h3>October 10, 2014</h3> | ||
| + | KOBAYASHI | ||
| + | <br> | ||
| + | <strong>GC Analysis</strong> | ||
| + | <br> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th rowspan="2">Host</th> | ||
| + | <th colspan="3">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>BY4247</td> | ||
| + | <td>p427-TEF</td> | ||
| + | <td>CuMTS3</td> | ||
| + | <td>pGEX6P-2F2, pGEX6P-2R</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | <img src="/wiki/images/b/b1/Kit_October10_c.jpg"> | ||
| + | <div class="clear"><hr /></div> | ||
| + | For More Information: <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1448000">BBa_K1448000</a> | ||
| + | </p> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
<div class="clear"><hr /></div> | <div class="clear"><hr /></div> | ||
| - | + | <div id='kitfooter-box' class='noprint'> | |
<div><a href="http://www.mediawiki.org/" style="float:left;margin:10px auto;"><img src="/wiki/skins/common/images/poweredby_mediawiki_88x31.png" height="31" width="88" alt="Powered by MediaWiki" /></a> | <div><a href="http://www.mediawiki.org/" style="float:left;margin:10px auto;"><img src="/wiki/skins/common/images/poweredby_mediawiki_88x31.png" height="31" width="88" alt="Powered by MediaWiki" /></a> | ||
| - | </div> | + | </div> |
| + | <div><a href="http://creativecommons.org/licenses/by/3.0/" style="float:right;margin:10px auto;z-index:0;"><img src="http://i.creativecommons.org/l/by/3.0/88x31.png" alt="Attribution 3.0 Unported" width="88" height="31" /></a> | ||
<div id="kitfooter"> | <div id="kitfooter"> | ||
<ul id="fkit"> | <ul id="fkit"> | ||
| Line 3,383: | Line 6,304: | ||
</ul> | </ul> | ||
</div> | </div> | ||
| - | + | ||
</div> | </div> | ||
</div> <!-- close kitfooter-box --> | </div> <!-- close kitfooter-box --> | ||
| Line 3,394: | Line 6,315: | ||
h3{ | h3{ | ||
border-bottom:2px dashed rgba(0,200,50,0.8); | border-bottom:2px dashed rgba(0,200,50,0.8); | ||
| + | } | ||
| + | |||
| + | th{ | ||
| + | border:1px solid rgba(300,300,300,300.5); | ||
| + | } | ||
| + | |||
| + | td{ | ||
| + | border:1px solid rgba(300,300,300,300.5); | ||
| + | } | ||
| + | |||
| + | th{ | ||
| + | padding:10px; | ||
| + | } | ||
| + | |||
| + | img{ | ||
| + | margin:10px 0px; | ||
} | } | ||
</style> | </style> | ||
Latest revision as of 01:13, 18 October 2014


LabNote
June
June 10, 2014
OKAMOTO
Miniprep
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| BL21(DE3) | pGEX6P-2 | CuMTSE1 |
| BL21(DE3) | pGEX6P-2 | CuMTSE2 |
| BL21(DE3) | pGEX6P-2 | CuMTS3 |
| BL21(DE3) | pGEX6P-2 | CuMTS62 |
Restriction Digest
| Host | Sample | Enzyme | Buffer | |
|---|---|---|---|---|
| Vector | Insert | |||
| BL21(DE3) | pGEX6P-2 | CuMTSE1 | EcoR1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTSE2 | EcoR1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTS3 | EcoR1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTS62 | EcoR1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTSE1 | Sal1, Not1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTSE2 | Sal1, Not1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTS3 | Sal1, Not1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTS62 | Sal1, Not1 | Buffer H |
June 11, 2014
KOBAYASHI
AGE
| Lane | Sample | Enzyme | ||
|---|---|---|---|---|
| Marker | Vector | Insert | ||
| 1 | λDNA-HindⅢ | |||
| 2 | pGEX6P-2 | CuMTSE1 | Non Treated | |
| 3 | pGEX6P-2 | CuMTSE1 | EcoR1 | |
| 4 | pGEX6P-2 | CuMTSE1 | Sal1, Not1 | |
| 5 | pGEX6P-2 | CuMTSE2 | Non Treated | |
| 6 | pGEX6P-2 | CuMTSE2 | EcoR1 | |
| 7 | pGEX6P-2 | CuMTSE2 | Sal1, Not1 | |
| 8 | pGEX6P-2 | CuMTS3 | Non Treated | |
| 9 | pGEX6P-2 | CuMTS3 | EcoR1 | |
| 10 | pGEX6P-2 | CuMTS3 | Sal1, Not1 | |
| 11 | pGEX6P-2 | CuMTS62 | Non Treated | |
| 12 | pGEX6P-2 | CuMTS62 | EcoR1 | |
| 13 | pGEX6P-2 | CuMTS62 | Sal1, Not1 | |

June 12, 2014
KOBAYASHI
Transformation
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS3 |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS62 |
June 13, 2014
KOBAYASHI
Colony Isolation
Select 6 colonies per plate from 4 plates from June 12.
| Host | Sample | Section | |
|---|---|---|---|
| Vector | Insert | ||
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | 1-a, 1-b |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | 2-a, 2-b |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS3 | 3-a, 3-b |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS62 | 6-a, 6-b |
June 14, 2014
ONISHI
Miniprep
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| 1-a | pGEX6P-2 | CuMTSE1 |
| 1-b | pGEX6P-2 | CuMTSE1 |
| 2-a | pGEX6P-2 | CuMTSE2 |
| 2-b | pGEX6P-2 | CuMTSE2 |
| 3-a | pGEX6P-2 | CuMTS3 |
| 3-b | pGEX6P-2 | CuMTS3 |
| 6-a | pGEX6P-2 | CuMTS62 |
| 6-b | pGEX6P-2 | CuMTS62 |
Miniprep
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| BL21(DE3) | pGEX6P-2 | CuMTSE1 |
| BL21(DE3) | pGEX6P-2 | CuMTSE2 |
| BL21(DE3) | pGEX6P-2 | CuMTS3 |
| BL21(DE3) | pGEX6P-2 | CuMTS62 |
Restriction Digest
| Host | Sample | Enzyme | Buffer | |
|---|---|---|---|---|
| Vector | Insert | |||
| 1-a | pGEX6P-2 | CuMTSE1 | EcoR1 | Buffer H |
| 1-b | pGEX6P-2 | CuMTSE1 | EcoR1 | Buffer H |
| 2-a | pGEX6P-2 | CuMTSE2 | EcoR1 | Buffer H |
| 2-b | pGEX6P-2 | CuMTSE2 | EcoR1 | Buffer H |
| 3-a | pGEX6P-2 | CuMTS3 | EcoR1 | Buffer H |
| 3-b | pGEX6P-2 | CuMTS3 | EcoR1 | Buffer H |
| 6-a | pGEX6P-2 | CuMTS62 | EcoR1 | Buffer H |
| 6-b | pGEX6P-2 | CuMTS62 | EcoR1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTSE1 | EcoR1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTSE2 | EcoR1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTS3 | EcoR1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTS62 | EcoR1 | Buffer H |
AGE
| Lane | Sample | Insert | Enzyme | Buffer | ||
|---|---|---|---|---|---|---|
| Marker | Host | Vector | ||||
| 1 | 1kb Ladder | |||||
| 2 | λDNA-HindⅢ | |||||
| 3 | 1-a | pGEX6P-2 | CuMTSE1 | EcoR1 | Buffer H | |
| 4 | 1-b | pGEX6P-2 | CuMTSE1 | EcoR1 | Buffer H | |
| 5 | 2-a | pGEX6P-2 | CuMTSE2 | EcoR1 | Buffer H | |
| 6 | 2-b | pGEX6P-2 | CuMTSE2 | EcoR1 | Buffer H | |
| 7 | 3-a | pGEX6P-2 | CuMTS3 | EcoR1 | Buffer H | |
| 8 | 3-b | pGEX6P-2 | CuMTS3 | EcoR1 | Buffer H | 9 | 6-a | pGEX6P-2 | CuMTS62 | EcoR1 | Buffer H |
| 10 | 6-b | pGEX6P-2 | CuMTS62 | EcoR1 | Buffer H | |
| 11 | BL21(DE3) | pGEX6P-2 | CuMTSE1 | EcoR1 | Buffer H | |
| 12 | BL21(DE3) | pGEX6P-2 | CuMTSE2 | EcoR1 | Buffer H | |
| 13 | BL21(DE3) | pGEX6P-2 | CuMTS3 | EcoR1 | Buffer H | |
| 14 | BL21(DE3) | pGEX6P-2 | CuMTS62 | EcoR1 | Buffer H | |
Apply λDNA-HindⅢ to compare and contrast AGE of July 14 and June 11.

June 16, 2014
YAMAJI
Preculture
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS3 |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS62 |
| BL21(DE3)pLysS | pGEX6P-2 | |
Main Culture
| Host | Sample | IPTG | |
|---|---|---|---|
| Vector | Insert | ||
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS3 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS62 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | Induced | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Non-induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Non-induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS3 | Non-induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS62 | Non-induced |
| BL21(DE3)pLysS | pGEX6P-2 | Non-induced | |
June 18, 2014
YAMAJI
Preculture
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 |
Main Culture
| Host | Sample | IPTG | |
|---|---|---|---|
| Vector | Insert | ||
| BL21(DE3)pLysS | pGEX6P-2 | Induced | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | Non-induced | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Non-induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Non-induced |
Protein Extraction (E. coli)
| Host | Sample | IPTG | |
|---|---|---|---|
| Vector | Insert | ||
| BL21(DE3)pLysS | pGEX6P-2 | Induced | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | Non-induced | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Non-induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Non-induced |
Preparations for SDS-PAGE
Add SDS sample buffer to protein extractions in a flask, incubate at 37℃ for 15 minutes and keep it at 4℃.

June 19, 2014
KITOMI
SDS-PAGE
| Host | Sample | IPTG | |
|---|---|---|---|
| Vector | Insert | ||
| BL21(DE3)pLysS | pGEX6P-2 | Induced | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | Non-induced | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Non-induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Non-induced |
June 21, 2014
SOGA
Miniprep
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 |
PCR
| Vector | Sample | DNA Polymerase | |
|---|---|---|---|
| Insert | Primer | ||
| pGEX6P-2 | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | KOD-FX-NEO |
AGE
| Lane | Sample | Insert | Primer | ||
|---|---|---|---|---|---|
| Marker | Host | Vector | |||
| 1 | 1kb Ladder | ||||
| 2 | BY4247 | p427-TEF | |||
| 3 | pGEX6P-2 | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | ||
| 4 | pGEX6P-2 | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | ||
| 5 | pGEX6P-2 | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | ||
| 6 | pGEX6P-2 | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | ||
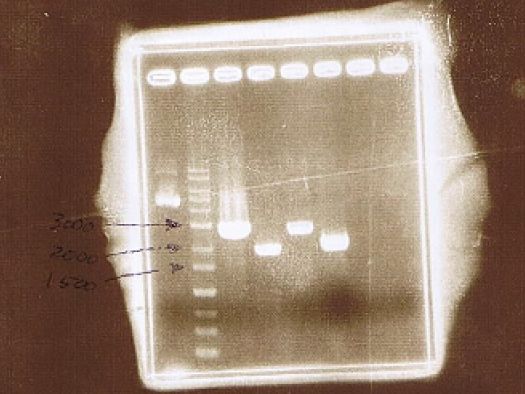
June 24, 2014
SOGA
Preculture
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| BL21(DE3)pLysS | ||
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 |
June 25, 2014
KOBAYASHI
Cultivate following samples in Big Scale at 30℃ overnight in shaken culture.
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| BL21(DE3)pLysS | ||
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 |
DNA Refinement 1
| Sample | |
|---|---|
| DNA | Primer |
| CuMTSE1 | pGEX6P-2F1, pGEX6P-2R |
| CuMTSE1 | pGEX6P-2F2, pGEX6P-2R |
Restriction Digest
| Vector | Sample | Enzyme | Buffer | |
|---|---|---|---|---|
| DNA | Primer | |||
| CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Buffer H | |
| CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Spe1, Xho1 | Buffer H | |
| p427-TEF | Spe1, Xho1 | Buffer H | ||
DNA Refinement 2
| Vector | Sample | Enzyme | |
|---|---|---|---|
| DNA | Primer | ||
| CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | |
| CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Spe1, Xho1 | |
| p427-TEF | Spe1, Xho1 | ||
Main Culture
| Host | Sample | IPTG | |
|---|---|---|---|
| Vector | Insert | ||
| BL21(DE3)pLysS | pGEX6P-2 | Induced | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | Non-induced | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Non-induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Non-induced |

Ligation
| Sample | ||
|---|---|---|
| Vector | DNA | Primer |
| p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R |
| p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R |
| p427-TEF | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R |
| p427-TEF | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R |
Transformation
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| DH5α | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R |
| DH5α | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R |
| DH5α | p427-TEF | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R |
| DH5α | p427-TEF | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R |
June 26, 2014
SAWANO
Colony Isolation
Isolate each colony from the plates which were inoculated on June 25.
| Host | Sample | Section | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| DH5α | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | 1~4 |
| DH5α | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | a~h |
| DH5α | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | ア~タ |
June 27, 2014
SHIMADA
Colony Sweep
| Lane | Sample | ||
|---|---|---|---|
| Host | Vector | Insert | |
| 1 | BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 |
| 2 | 1 | ||
| 3 | 2 | ||
| 4 | 3 | ||
| 5 | 4 | ||
| 6 | a | ||
| 7 | b | ||
| 8 | c | ||
| 9 | d | ||
| 10 | e | ||
| 11 | f | ||
| 12 | g | ||
| 13 | h | ||

Colony Sweep
| Lane | Sample | ||
|---|---|---|---|
| Host | Vector | Insert | |
| 1 | BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 |
| 2 | ア | ||
| 3 | イ | ||
| 4 | ウ | ||
| 5 | エ | ||
| 6 | オ | ||
| 7 | カ | ||
| 8 | キ | ||
| 9 | ク | ||
| 10 | ケ | ||
| 11 | コ | ||
| 12 | サ | ||
| 13 | シ | ||
| 14 | ス | ||
| 15 | セ | ||
| 16 | ソ | ||
| 17 | タ | ||

Identified targeted genes in 4, c, h, イ, カ, セ
Miniprep
| Sample | |
|---|---|
| Host | |
| 4 | |
| C | |
| H | |
| イ | |
| カ | |
| セ | |
Restriction Digest
| Sample | Enzyme | Buffer |
|---|---|---|
| Host | ||
| 4 | Spe1, Xho1 | Buffer H |
| C | Spe1, Xho1 | Buffer H |
| H | Spe1, Xho1 | Buffer H |
| イ | Spe1, Xho1 | Buffer H |
| カ | Spe1, Xho1 | Buffer H |
AGE
| Lane | Sample | Primer | Enzyme | ||
|---|---|---|---|---|---|
| Marker | Host | Insert | |||
| 1 | 1kb Ladder | ||||
| 2 | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | |||
| 3 | 4 | Spe1, Xho1 | |||
| 4 | 4 | Spe1, Xho1 | |||
| 5 | 4 | Non Treated | |||
| 6 | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | |||
| 7 | c | Spe1, Xho1 | |||
| 8 | c | Spe1, Xho1 | |||
| 9 | c | Non Treated | |||
| 10 | h | Spe1, Xho1 | |||
| 11 | h | Spe1, Xho1 | |||
| 12 | h | Non Treated | |||

Transformation (electroportation)
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| BY4247 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R |
| BY4247 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R |
June 30, 2014
SOGA
Inoculation
Divide each of 2 plates into 8 sections and inoculate following samples on them.
| Host | Sample | Section | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| BY4247 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | A~H |
| BY4247 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | 一~ハ |
July
July 1, 2014
KOBAYASHI
Protein Extraction (S. cerevisiae)
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| BY4247 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R |
| BY4247 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R |
July 2, 2014
ONISHI
Miniprep
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| BY4247 | p427-TEF | ||
| BY4247 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R |
| BY4247 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R |
Restriction Digest
| Sample | Enzyme | Buffer | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| p427-TEF | CuMTSE1 | EcoR1 | Buffer H | |
| p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | EcoR1 | Buffer H |
| p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | EcoR1 | Buffer H |
| p427-TEF | CuMTSE1 | Sac1, Kpn1 | Buffer L | |
| p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Sac1, Kpn1 | Buffer L |
| p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Sac1, Kpn1 | Buffer L |
AGE
| Lane | Sample | Enzyme | |||
|---|---|---|---|---|---|
| Marker | Vector | Insert | Primer | ||
| 1 | p427-TEF | Non Treated | |||
| 2 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Non Treated | |
| 3 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Non Treated | |
| 4 | 1kb Ladder | ||||
| 5 | p427-TEF | EcoR1 | |||
| 6 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | EcoR1 | |
| 7 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | EcoR1 | |
| 8 | 1kb Ladder | ||||
| 9 | p427-TEF | Kpn1 | |||
| 10 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Kpn1 | |
| 11 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Kpn1 | |
| 12 | 1kb Ladder | ||||
| 13 | p427-TEF | Sac1 | |||
| 14 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Sac1 | |
| 15 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Sac1 | |

Restriction Digest
| Sample | Enzyme | Buffer | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| p427-TEF | CuMTSE1 | EcoR1 | Buffer H | |
| p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | EcoR1 | Buffer H |
| p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | EcoR1 | Buffer H |
| p427-TEF | CuMTSE1 | Sac1 | Buffer L | |
| p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Sac1 | Buffer L |
| p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Sac1 | Buffer L |
| p427-TEF | CuMTSE1 | Kpn1 | Buffer L | |
| p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Kpn1 | Buffer L |
| p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Kpn1 | Buffer L |
| p427-TEF | CuMTSE1 | Spe1, Xho1 | Buffer H | |
| p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Buffer H |
| p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Spe1, Xho1 | Buffer H |
AGE
| Lane | Sample | Enzyme | |||
|---|---|---|---|---|---|
| Marker | Vector | Insert | Primer | ||
| 1 | 1kb Ladder | ||||
| 2 | p427-TEF | CuMTSE1 | EcoR1 | ||
| 3 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | EcoR1 | |
| 4 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | EcoR1 | |
| 5 | p427-TEF | CuMTSE1 | Kpn1 | ||
| 6 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Kpn1 | |
| 7 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Kpn1 | |
| 8 | p427-TEF | CuMTSE1 | Sac1 | ||
| 9 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Sac1 | |
| 10 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Sac1 | |
| 11 | p427-TEF | CuMTSE1 | Spe1, Xho1 | ||
| 12 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | |
| 13 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Spe1, Xho1 | |
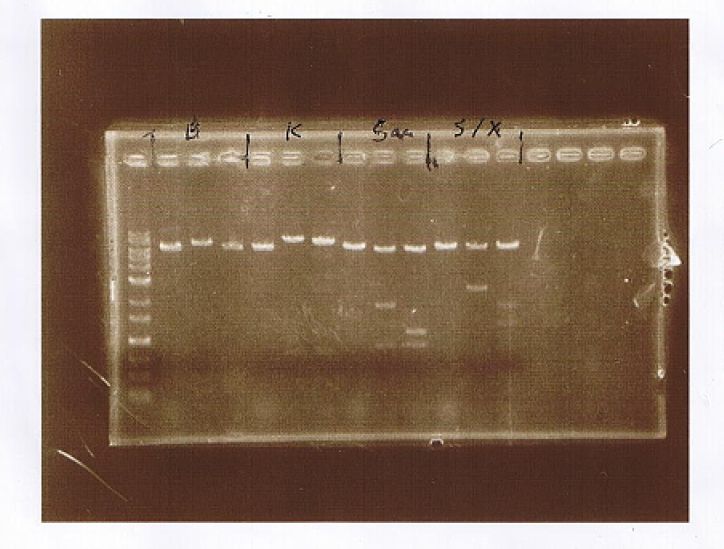
July 3, 2014
OKAMOTO
PCR
| Sample | DNA Polymerase | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| pGEX6P-2 | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTS3 | pGEX6P-2F2, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTS62 | pGEX6P-2F2, pGEX6P-2R | KOD-FX-NEO |
Protein Extraction (S. cerevisiae)
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| BY4247 | p427-TEF | ||
| A | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R |
| 六 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R |
July 4, 2014
SHIMADA
AGE
| Lane | Sample/Marker | |||
|---|---|---|---|---|
| Marker | Vector | Insert | Primer | |
| 1 | 1kb Ladder | |||
| 2 | pGEX6P-2 | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | |
| 3 | pGEX6P-2 | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | |
| 4 | pGEX6P-2 | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | |
| 5 | pGEX6P-2 | CuMTS3 | pGEX6P-2F2, pGEX6P-2R | |
| 6 | pGEX6P-2 | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | |
| 7 | pGEX6P-2 | CuMTS62 | pGEX6P-2F2, pGEX6P-2R | |

PCR
| Sample | DNA Polymerase | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| pGEX6P-2 | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTS3 | pGEX6P-2F2, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTS62 | pGEX6P-2F2, pGEX6P-2R | KOD-FX-NEO |
AGE
| Lane | Sample | |||
|---|---|---|---|---|
| Marker | Vector | Insert | Primer | |
| 1 | 1kb Ladder | |||
| 2 | pGEX6P-2 | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | |
| 3 | pGEX6P-2 | 1kb Ladder+K83:N91 | pGEX6P-2F2, pGEX6P-2R | |
| 4 | pGEX6P-2 | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | |
| 5 | pGEX6P-2 | CuMTS3 | pGEX6P-2F2, pGEX6P-2R | |
| 6 | pGEX6P-2 | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | |
| 7 | pGEX6P-2 | CuMTS62 | pGEX6P-2F2, pGEX6P-2R | |

July 5, 2014
KOBAYASHI
SDS-PAGE
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| BL21(DE3)pLysS | pGEX6P-2 | ||
| 4 | pGEX6P-2 | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R |
| C | pGEX6P-2 | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R |
| BY4247 | p427-TEF | ||
| A | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R |
| 六 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R |
July 6, 2014
SOGA
DNA Refinement 1
| Sample | |
|---|---|
| DNA | Primer |
| CuMTSE2 | pGEX6P-2F1, pGEX6P-2R |
| CuMTSE2 | pGEX6P-2F2, pGEX6P-2R |
| CuMTS3 | pGEX6P-2F1, pGEX6P-2R |
| CuMTS62 | pGEX6P-2F1, pGEX6P-2R |
Restriction Digest
| Vector | Sample | Enzyme | Buffer | |
|---|---|---|---|---|
| DNA | Primer | |||
| CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer | |
| CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer | |
| CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer | p427-TEF | Spe1, Xho1 | Fast digest buffer |
DNA Refinement 2
| Sample | Enzyme | |
|---|---|---|
| DNA | Primer | |
| CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
| CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | Spe1, Xho1 |
Ligation
| Vector | Sample | Enzyme | |
|---|---|---|---|
| DNA | Primer | ||
| p427-TEF | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
| p427-TEF | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | Spe1, Xho1 |
DNA ligase: DNA Ligation Kit ‹Mighty Mix› (TAKARA BIO INC.)
Transformation (E. coli)
| Host | Sample | Enzyme | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| DH5α | p427-TEF | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
| DH5α | p427-TEF | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | Spe1, Xho1 |
| DH5α | p427-TEF | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
| DH5α | p427-TEF | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
July 7, 2014
KOBAYASHI
Colony Isolation
Isolate each colony from the plates which were inoculated on July 6.
| Host | Sample | Section | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| DH5α | p427-TEF | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | 1~16 |
| DH5α | p427-TEF | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | ア~エ |
| DH5α | p427-TEF | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | a~p |
Transformation (E. coli)
| Host | Sample | Enzyme | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| DH5α | p427-TEF | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | Spe1, Xho1 |
July 8, 2014
OKAMOTO
Colony Sweep
| Lane | Sample | |
|---|---|---|
| Host | ||
| 1 | DH5α | p427-TEF |
| 2 | a | |
| 3 | b | |
| 4 | c | |
| 5 | d | |
| 6 | e | |
| 7 | f | |
| 8 | g | |
| 9 | h | |
| 10 | i | |
| 11 | j | |
| 12 | k | |
| 13 | l | |
| 14 | m | |
| 15 | n | |
| 16 | o | |
| 17 | p | |

Colony Sweep
| Lane | Sample | |
|---|---|---|
| Host | ||
| 1 | DH5α | p427-TEF |
| 2 | ア | |
| 3 | イ | |
| 4 | ウ | |
| 5 | エ | |

Colony Sweep
| Lane | Sample | |
|---|---|---|
| Host | ||
| 1 | DH5α | p427-TEF |
| 2 | 1 | |
| 3 | 2 | |
| 4 | 3 | |
| 5 | 4 | |
| 6 | 5 | |
| 7 | 6 | |
| 8 | 7 | |
| 9 | 8 | |
| 10 | 9 | |
| 11 | 10 | |
| 12 | 11 | |
| 13 | 12 | |
| 14 | 13 | |
| 15 | 14 | |
| 16 | 15 | |
| 17 | 16 | |

July 9, 2014
OKAMOTO
Main Culture
| Sample |
|---|
| Host |
| d |
| h |
| l |
| ア |
| イ |
| エ |
Mini Prep
| Sample |
|---|
| Host |
| D |
| H |
| L |
| ア |
| イ |
| エ |
Restriction Digest
| Vector | Sample | Enzyme | Buffer | |
|---|---|---|---|---|
| Insert | Primer | |||
| p427-TEF(d) | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer |
| p427-TEF(h) | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer |
| p427-TEF(l) | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer |
| p427-TEF(ア) | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer |
| p427-TEF(イ) | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer |
| p427-TEF(エ) | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer |
AGE
| Lane | Sample | Enzyme | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| 1 | p427-TEF(ア) | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Non Treated |
| 2 | p427-TEF(ア) | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
| 3 | p427-TEF(イ) | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Non Treated |
| 4 | p427-TEF(イ) | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
| 5 | p427-TEF(エ) | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Non Treated |
| 6 | p427-TEF(エ) | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
| 7 | p427-TEF(d) | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Non Treated |
| 8 | p427-TEF(d) | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
| 9 | p427-TEF(h) | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Non Treated |
| 10 | p427-TEF(h) | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
| 11 | p427-TEF(l) | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Non Treated |
| 12 | p427-TEF(l) | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |

August
August 19, 2014
KOBAYASHI
Conduct mutagenesis to remove an illegal restriction site (EcoR1)
InversePCR
| Sample | |||
|---|---|---|---|
| Vector | Insert | Primer | DNA polymerase |
| pGEX6P-2 | CuMTS3 | Terpinene3-mutagenesis-FP, Terpinene3-mutagenesis-RP | KOD-Plus |
August 20, 2014
KOBAYASHI
Restart mutagenesis from KOD -Plus- Mutagenesis Kit protocol No.3 to obtain plasmid (muta-CuMTS3 in pGEX-6P-2)
Transformation
| Sample | |||
|---|---|---|---|
| Host | Vector | Insert | Primer |
| DH5α | pGEX6P-2 | muta-CuMTS3 | Terpinene3-mutagenesis-FP and Terpinene3-mutagenesis-RP |
August 21, 2014
KOBAYASHI
Colony Isolation
Isolate each colony from two plates which were inoculated overnight
| Sample | Section | |||
|---|---|---|---|---|
| Host | Vector | Insert | Primer | |
| DH5α | pSB1C3 | muta-CuMTS3 | Terpinene3-mutagenesis-FP, Terpinene3-mutagenesis-RP |
あ-み |
August 22, 2014
KOBAYASHI
Restriction Digest
| Sample | Enzyme | Buffer | ||
|---|---|---|---|---|
| Host | Vector | Insert | ||
| DH5α あ | pGEX6P-2 | muta-CuMTS3 | Sal1 | Fast Digest buffer |
| DH5α か | pGEX6P-2 | muta-CuMTS3 | Sal1 | Fast Digest buffer |
| DH5α さ | pGEX6P-2 | muta-CuMTS3 | Sal1 | Fast Digest buffer |
| DH5α た | pGEX6P-2 | muta-CuMTS3 | Sal1 | Fast Digest buffer |
AGE
| Lane | Sample | Enzyme | |||
|---|---|---|---|---|---|
| Marker | Host | Vector | Insert | ||
| 1 | 1kb Ladder | ||||
| 2 | DH5α あ | pGEX6P-2 | muta-CuMTS3 | Sal1 | |
| 3 | DH5α か | pGEX6P-2 | muta-CuMTS3 | Sal1 | |
| 4 | DH5α さ | pGEX6P-2 | muta-CuMTS3 | Sal1 | |
| 5 | DH5α た | pGEX6P-2 | muta-CuMTS3 | Sal1 | |
Restriction Digest
| Sample | Enzyme | Buffer | ||
|---|---|---|---|---|
| Host | Vector | Insert | ||
| DH5α か | pGEX6P-2 | CuMTS3 | EcoR1 | Fast Digest buffer |
| DH5α か | pGEX6P-2 | muta-CuMTS3 | EcoR1 | Fast Digest buffer |
AGE
| Lane | Sample | Enzyme | |||
|---|---|---|---|---|---|
| Marker | Host | Vector | Insert | ||
| 1 | 1kb Ladder | ||||
| 2 | DH5α か | pGEX6P-2 | CuMTS3 | EcoR1 | |
| 3 | DH5α か | pGEX6P-2 | muta-CuMTS3 | EcoR1 | |
 An illegal EcoR1 restriction site was removed.
An illegal EcoR1 restriction site was removed.PCR
| Sample | DNA polymerase | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| か pGEX6P-2 | muta-CuMTS3 | terpinene3-prefix and terpinene3-suffix | KOD-FX-NEO-Plus |
AGE
| Lane | Sample | DNA polymerase | |||
|---|---|---|---|---|---|
| Marker | Vector | Insert | Primer | ||
| 1 | 1kb Ladder | ||||
| 2 | か pGEX6P-2 | muta-CuMTS3 | terpinene3-prefix and terpinene3-suffix | KOD-FX-NEO-Plus | |

August 30, 2014
KOBAYASHIPCR
| Sample | DNA polymerase | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| か pGEX6P-2 | muta-CuMTS3 | terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1 | Taq |
AGE
| Lane | Sample | |||
|---|---|---|---|---|
| Marker | Vector | Insert | Primer | |
| 1 | 1kb Ladder | |||
| 2 | か pGEX6P-2 | muta-CuMTS3 | terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1 | |
 Made total amount of 295 μL of PCR samples.
Made total amount of 295 μL of PCR samples.
September
September 18, 2014
KOBAYASHIMiniprep
| Sample | ||
|---|---|---|
| Host | Vector | Insert |
| DH5α | pSB1C3 | BBa_J04450 |
| DH5α | pSB1C3 | BBa_J04450 |
| DH5α | pSB1C3 | BBa_J04450 |
Restriction Digest
| Sample | Buffer | ||
|---|---|---|---|
| Vector | Insert | Enzyme | |
| pSB1C3 | BBa_J04450 | EcoR1, Pst1 | Buffer H |
| pSB1C3 | BBa_J04450 | Xba1, Spe1 | Fast Digest buffer |
| pSB1C3 | BBa_J04450 | ||
AGE
| Lane | Sample | Enzyme | |||
|---|---|---|---|---|---|
| Marker | Host | Vector | Insert | ||
| 1 | 1kb Ladder | ||||
| 2 | DH5α | pSB1C3 | BBa_J04450 | EcoR1, Pst1 | |
| 3 | DH5α | pSB1C3 | BBa_J04450 | Xba1, Spe1 | |
| 4 | DH5α | pSB1C3 | BBa_J04450 | ||

PCR
| Sample | DNA polymerase | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| か pGEX6P-2 | muta-CuMTS3 | terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1 | Taq |
DNA Refinement 1
| Sample | |
|---|---|
| Insert | Primer |
| muta-CuMTS3 | terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1 |
Restriction Digest
| Sample | Enzyme | Buffer | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| pSB1C3 | Xba1, Spe1 | Fast Digest | ||
| か pGEX6P-2 | muta-CuMTS3 | terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1 | Xba1, Spe1 | Fast Digest |
DNA Refinement 2
| Sample | |
|---|---|
| Insert | Primer |
| muta-CuMTS3 | terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1 |
Ligation
| Sample | Enzyme | ||
|---|---|---|---|
| Vector | DNA | Primer | |
| pSB1C3 | muta-CuMTS3 | terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1 | Spe1, Xba1 |
Transformation
| Sample | |||
|---|---|---|---|
| Host | Vector | Insert | Primer |
| DH5α | pSB1C3 | muta-CuMTS3 | terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1 |
September 19, 2014
KOBAYASHIColony Isolation
Isolate each colony from the plates which were inoculated overnight.
| Sample | Section | |||
|---|---|---|---|---|
| Host | Vector | Insert | Primer | |
| DH5α | pSB1C3 | muta-CuMTS3 | terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1 | A-P |
| DH5α | pSB1C3 | muta-CuMTS3 | terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1 | ア-タ |
Colony Sweep
| Lane | Sample | ||
|---|---|---|---|
| Host | Vector | Insert | |
| 1 | DH5α | pSB1C3 | muta-CuMTS3 |
| 2 | ア | ||
| 3 | イ | ||
| 4 | ウ | ||
| 5 | エ | ||
| 6 | オ | ||
| 7 | カ | ||
| 8 | キ | ||
| 9 | ク | ||
| 10 | ケ | ||
| 11 | コ | ||
| 12 | サ | ||
| 13 | シ | ||
| 14 | ス | ||
| 15 | セ | ||
| 16 | ソ | ||
| 17 | タ | ||
Targeted colonies were identified in セ and ソ.
Colony Sweep
| Lane | Sample | ||
|---|---|---|---|
| Host | Vector | Insert | |
| 1 | DH5α | pSB1C3 | muta-CuMTS3 |
| 2 | A | ||
| 3 | B | ||
| 4 | C | ||
| 5 | D | ||
| 6 | E | ||
| 7 | F | ||
| 8 | G | ||
| 9 | H | ||
| 10 | I | ||
| 11 | J | ||
| 12 | K | ||
| 13 | L | ||
| 14 | M | ||
| 15 | N | ||
| 16 | O | ||
| 17 | P | ||

Targeted colonies were identified in colony N and F.
Select セ, ソ, N, F
Main Culture
| Sample | |||
|---|---|---|---|
| Host | Vector | Insert | Primer |
| DH5α(N) | pSB1C3 | muta-CuMTS3 | ---------- |
| DH5α(F) | pSB1C3 | muta-CuMTS3 | ---------- |
| DH5α(セ) | pSB1C3 | muta-CuMTS3 | ---------- |
| DH5α(ソ) | pSB1C3 | muta-CuMTS3 | ---------- |
Cultivate these samples in 3mL of LB medium with ChlPhe at 30°C overnight.
September 20, 2014
KOBAYASHIMiniprep
| Sample | ||
|---|---|---|
| Host | Vector | Primer |
| DH5α(N) | pSB1C3 | muta-CuMTS3 |
| DH5α(F) | pSB1C3 | muta-CuMTS3 |
| DH5α(セ) | pSB1C3 | muta-CuMTS3 |
| DH5α(ソ) | pSB1C3 | muta-CuMTS3 |
Enzyme: E/P and Xba1
Restriction Digest
| Sample | Enzyme | Buffer | |||
|---|---|---|---|---|---|
| Marker | Host | Vector | Insert | ||
| pSB1C3 | |||||
| DH5α (セ) | pSB1C3 | muta-CuMTS3 | |||
| DH5α (ソ) | pSB1C3 | muta-CuMTS3 | |||
| DH5α (F) | pSB1C3 | muta-CuMTS3 | |||
| DH5α (N) | pSB1C3 | muta-CuMTS3 | |||
| DH5α (セ) | pSB1C3 | EcoR1, Pst1 | Buffer H | ||
| DH5α (ソ) | pSB1C3 | muta-CuMTS3 | EcoR1, Pst1 | Buffer H | |
| DH5α (F) | pSB1C3 | muta-CuMTS3 | EcoR1, Pst1 | Buffer H | |
| DH5α (N) | pSB1C3 | muta-CuMTS3 | EcoR1, Pst1 | Buffer H | |
| pSB1C3 | muta-CuMTS3 | EcoR1, Pst1 | Buffer H | ||
| pSB1C3 | Xba1 | FD buffer | |||
| DH5α (セ) | pSB1C3 | muta-CuMTS3 | Xba1 | FD buffer | |
| DH5α (ソ) | pSB1C3 | muta-CuMTS3 | Xba1 | FD buffer | |
| DH5α (F) | pSB1C3 | muta-CuMTS3 | Xba1 | FD buffer | |
| DH5α (N) | pSB1C3 | muta-CuMTS3 | Xba1 | FD buffer | |
AGE
| Lane | Sample | Enzyme | |||
|---|---|---|---|---|---|
| Marker | Host | Vector | Insert | ||
| 1 | pSB1C3 | ||||
| 2 | DH5α (セ) | pSB1C3 | muta-CuMTS3 | ||
| 3 | DH5α (ソ) | pSB1C3 | muta-CuMTS3 | ||
| 4 | DH5α (F) | pSB1C3 | muta-CuMTS3 | ||
| 5 | DH5α (N) | pSB1C3 | muta-CuMTS3 | ||
| 6 | 1kb Ladder | ||||
| 7 | DH5α (セ) | pSB1C3 | muta-CuMTS3 | EcoR1, Pst1 | |
| 8 | DH5α (ソ) | pSB1C3 | muta-CuMTS3 | EcoR1, Pst1 | |
| 9 | DH5α (F) | pSB1C3 | muta-CuMTS3 | EcoR1, Pst1 | |
| 10 | DH5α (N) | pSB1C3 | muta-CuMTS3 | EcoR1, Pst1 | |
| 11 | pSB1C3 | muta-CuMTS3 | EcoR1, Pst1 | ||
| 12 | 1kb Ladder | ||||
| 13 | pSB1C3 | Xba1 | |||
| 14 | DH5α (セ) | pSB1C3 | muta-CuMTS3 | Xba1 | |
| 15 | DH5α (ソ) | pSB1C3 | muta-CuMTS3 | Xba1 | |
| 16 | DH5α (F) | pSB1C3 | muta-CuMTS3 | Xba1 | |
| 17 | DH5α (N) | pSB1C3 | muta-CuMTS3 | Xba1 | |

Main Culture
| Sample | |||
|---|---|---|---|
| Host | Vector | Insert | Primer |
| DH5α(F) | pSB1C3 | muta-CuMTS3 | ---------- |
| DH5α(F) | pSB1C3 | muta-CuMTS3 | ---------- |
| DH5α(F) | pSB1C3 | muta-CuMTS3 | ---------- |
Cultivate these samples in 3mL of LB medium with ChlPhe at 30°C overnight.
September 21, 2014
KOBAYASHIMiniprep
| Sample | |||
|---|---|---|---|
| Host | Vector | Insert | Primer |
| DH5α(F) | pSB1C3 | muta-CuMTS3 | |
| DH5α(F) | pSB1C3 | muta-CuMTS3 | |
| DH5α(F) | pSB1C3 | muta-CuMTS3 | |
Restriction Digest
| Sample | Enzyme | Buffer | ||||
|---|---|---|---|---|---|---|
| Marker | Host | Vector | Insert | Primer | ||
| 1kb Ladder | ||||||
| pSB1C3 | BBa_J04450 | EcoR1 | Buffer H | |||
| pSB1C3 | BBa_J04450 | Pst1 | Buffer H | |||
| pSB1C3 | BBa_J04450 | EcoR1, Pst1 | Buffer H | |||
| pSB1C3 | BBa_J04450 | Spe1 | FD buffer | |||
| pSB1C3 | BBa_J04450 | Xba1 | FD buffer | |||
| pSB1C3 | BBa_J04450 | Spe1, Xba1 | FD buffer | |||
| pSB1C3 | BBa_J04450 | |||||
| λDNA-HindⅢ | ||||||
| F | pSB1C3 | muta-CuMTS3 | EcoR1 | Buffer H | ||
| F | pSB1C3 | muta-CuMTS3 | Pst1 | Buffer H | ||
| F | pSB1C3 | muta-CuMTS3 | EcoR1, Pst1 | Buffer H | ||
| F | pSB1C3 | muta-CuMTS3 | Spe1 | FD buffer | ||
| F | pSB1C3 | muta-CuMTS3 | Xba1 | FD buffer | ||
| F | pSB1C3 | muta-CuMTS3 | Spe1, Xba1 | FD buffer | ||
| F | pSB1C3 | muta-CuMTS3 | ||||
AGE
| Lane | Sample | Enzyme | ||||
|---|---|---|---|---|---|---|
| Marker | Host | Vector | Insert | Primer | ||
| 1 | 1kb Ladder | |||||
| 2 | pSB1C3 | BBa_J04450 | EcoR1 | |||
| 3 | pSB1C3 | BBa_J04450 | Pst1 | |||
| 4 | pSB1C3 | BBa_J04450 | EcoR1, Pst1 | |||
| 5 | pSB1C3 | BBa_J04450 | Spe1 | |||
| 6 | pSB1C3 | BBa_J04450 | Xba1 | |||
| 7 | pSB1C3 | BBa_J04450 | Spe1, Xba1 | |||
| 8 | pSB1C3 | BBa_J04450 | ||||
| 9 | λDNA-HindⅢ | |||||
| 10 | F | pSB1C3 | muta-CuMTS3 | EcoR1 | ||
| 11 | F | pSB1C3 | muta-CuMTS3 | Pst1 | ||
| 12 | F | pSB1C3 | muta-CuMTS3 | EcoR1, Pst1 | ||
| 13 | F | pSB1C3 | muta-CuMTS3 | Spe1 | ||
| 14 | F | pSB1C3 | muta-CuMTS3 | Xba1 | ||
| 15 | F | pSB1C3 | muta-CuMTS3 | Spe1, Xba1 | ||
| 16 | F | pSB1C3 | muta-CuMTS3 | |||
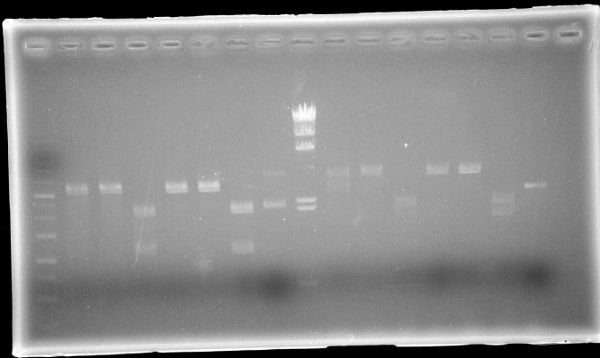
October
October 1, 2014
KOBAYASHIProtein Extraction (S. cerevisiae)
| Sample | |||
|---|---|---|---|
| Host | Vector | Insert | Primer |
| BY4247 | p427-TEF | ||
| BY4247 | p427-TEF | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R |
| BY4247 | p427-TEF | CuMTS3 | pGEX6P-2F2, pGEX6P-2R |
| BY4247 | p427-TEF | CuMTS62 | pGEX6P-2F2, pGEX6P-2R |
SDS-PAGE
| Lane | Sample | |||
|---|---|---|---|---|
| Host | Vector | Insert | Primer | |
| 1 | BY4247 | p427-TEF | ||
| 2 | BY4247 | p427-TEF | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R |
| 3 | BY4247 | p427-TEF | CuMTS3 | pGEX6P-2F2, pGEX6P-2R |
| 4 | BY4247 | p427-TEF | CuMTS62 | pGEX6P-2F2, pGEX6P-2R |
October 2, 2014
KOBAYASHIPreculture
| Sample | |||
|---|---|---|---|
| Host | Vector | Insert | Primer |
| BY4247 | p427-TEF | ||
| BY4247 | p427-TEF | CuMTS3 | pGEX6P-2F2, pGEX6P-2R |
Cultivate these samples in 3mL of SD medium with G418 at 30°C overnight.
Western blot
| Lane | Sample | |||
|---|---|---|---|---|
| Host | Vector | Insert | Primer | |
| 1 | BY4247 | p427-TEF | ||
| 2 | BY4247 | p427-TEF | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R |
| 3 | BY4247 | p427-TEF | CuMTS3 | pGEX6P-2F2, pGEX6P-2R |
| 4 | BY4247 | p427-TEF | CuMTS62 | pGEX6P-2F2, pGEX6P-2R |
October 3, 2014
KOBAYASHIMain culture
Cultivate following samples in Big Scale with G418 at 28°C overnight in shaken culture.
| Sample | |||
|---|---|---|---|
| Host | Vector | Insert | Primer |
| BY4247 | p427-TEF | ||
| BY4247 | p427-TEF | CuMTS3 | pGEX6P-2F2, pGEX6P-2R |
Restriction Digest
| Sample | Enzyme | Buffer | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| pSB1C3 | EcoR1, Spe1 | Buffer H | ||
| pSB1C3 | Xba1, Pst1 | Buffer H | ||
| Linearized pSB1A3 | EcoR1, Pst1 | Buffer H | ||
AGE
| Lane | Marker | Sample | Enzyme | ||
|---|---|---|---|---|---|
| Vector | Insert | Primer | |||
| 1 | 1kb Ladder | ||||
| 2 | pSB1C3 | EcoR1, Spe1 | |||
| 3 | pSB1C3 | Xba1, Pst1 | |||
| 4 | Linearized pSB1A3 | EcoR1, Pst1 | |||
DNA Refinement 1
| Sample | Enzyme | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| pSB1C3 | EcoR1, Spe1 | ||
| pSB1C3 | Xba1, Pst1 | ||
| pSB1C3 | Xba1, Pst1 | ||
| Linearized pSB1A3 | EcoR1, Pst1, Dpn1 | ||
Ligation
| Vector | Sample | Enzyme | |
|---|---|---|---|
| Insert1 | Insert2 | ||
| pSB1A3 | EcoR1, Xba1, Spe1, Pst1 | ||
| pSB1A3 | EcoR1, Xba1, Spe1, Pst1 | ||
DNA ligase: DNA Ligation Kit ‹Mighty Mix› (TAKARA BIO INC.)
Transformation
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert1 | Insert2 | |
| DH5α | pSB1A3 | ||
| DH5α | pSB1A3 | ||
October 4, 2014
KOBAYASHIColony Isolation
Isolate each colony from the plates which were inoculated overnight.
| Host | Sample | Section | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| DH5α | pSB1A3 | 1-16 | ||
| DH5α | pSB1A3 | a-p | ||
Monoterpene Extraction
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| BY4247 | p427-TEF | ||
| BY4247 | p427-TEF | CuMTS3 | pGEX6P-2F2, pGEX6P-2R |
Samples were stocked in -4℃.
Preculture
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| BY4247 | p427-TEF | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R |
| BY4247 | p427-TEF | CuMTS62 | pGEX6P-2F2, pGEX6P-2R |
Cultivate samples in 3mL of SD medium with G418 at 30°C overnight.
October 5, 2014
KOBAYASHIMiniprep
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| DH5α | pSB1A3 | |
| DH5α | pSB1A3 | |
Restriction Digest and AGE
| Host | Sample | Enzyme | |
|---|---|---|---|
| Vector | Insert | ||
| DH5α | pSB1C3 | EcoR1, Pst1 | |
| DH5α | pSB1C3 | EcoR1, Pst1 | |
| pSB1C3 | K1448000 | EcoR1, Pst1 | |
October 8, 2014
KOBAYASHIGC-MS analysis
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| BY4247 | p427-TEF | ||
| BY4247 | p427-TEF | CuMTS3 | pGEX6P-2F2, pGEX6P-2R |
Preculture
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| BY4247 | p427-TEF | ||
| BY4247 | p427-TEF | CuMTS3 | pGEX6P-2F2, pGEX6P-2R |
October 9, 2014
KOBAYASHIMain Culture
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| BY4247 | p427-TEF | ||
| BY4247 | p427-TEF | CuMTS3 | pGEX6P-2F2, pGEX6P-2R |
October 10, 2014
KOBAYASHIGC Analysis
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| BY4247 | p427-TEF | ||
| BY4247 | p427-TEF | CuMTS3 | pGEX6P-2F2, pGEX6P-2R |
 "
"
