Team:Evry/Interlab Study
From 2014.igem.org
| Line 4: | Line 4: | ||
<html> | <html> | ||
| + | </section> | ||
| + | </div> | ||
<script src="http://codyhouse.co/demo/vertical-timeline/js/modernizr.js"></script> | <script src="http://codyhouse.co/demo/vertical-timeline/js/modernizr.js"></script> | ||
<script src="https://2014.igem.org/Team:Evry/JS/Jqueryui?action=raw&ctype=text/javascript"></script> | <script src="https://2014.igem.org/Team:Evry/JS/Jqueryui?action=raw&ctype=text/javascript"></script> | ||
Revision as of 14:44, 23 August 2014
Interlab study

Aug 24

Contruction n°1: PSB1C3 with I20260
Digestion of PSB1C3 and PSB1K3-I20260 by EcoRI and PstI
- 1 µl EcoRI-HF
- 1 µl PstI
- 2 µl buffer
- 0.2 µl BSA
- 13.7 or 76 µl respectively for I20260 and PSB1C3
- 10.8 µl miliQ watter
Incubation 45 minutes at 37°C. Denaturation step 20 minutes at 80 °C.
Loading on a 1% agarose gel to perform an extraction from gel
Contruction n°2: PSB1C3 with J23101-E1010
Aug 23

Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
Transformation plate observation:
- BBa_J23101: 11 isolated colonies
- BBa_K823012: 100-150 isolated colonies
A PCR was performed with 8 colonies for BBa_K823012 and for BBa_J23101 following the protocol Table 3 and 4.
To verify PCR products, 10 µl was loaded on a 1% agarose gel with 2 µl of loading dye 6X.
We expected to obtain a band between 300 and 400 bp. So we decided to amplify the colony n°8. The colony was seed in 5 ml of LB medium and 5 µl of chloramphenicol. Culture was incubated overnight at 37°C with 200 rpm agitation.
We expected to obtain a band around 1200 bp. So we decided to amplify the colony n°2 corresponding to lane 11. The colony was seed in 5 ml of LB medium and 5 µl of chloramphenicol. Culture was incubated overnight at 37°C with 200 rpm agitation. Aug 22

Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
Transformations of registry parts (BBa_K823012 and J23101) were performed to obtain colonies with plasmid containing parts for future amplifications.
The transformation was performed on DH5 alpha E. coli, as followed:
- Remove E. coli competent tubes from -80°C and keep it on ice
- Add 3 µl of template (here solubilized plasmids from the registry distribution kit) and mix gently
- Incubate 10 minutes on ice
- Perform an heat shock 30 seconds at 42°C
- Incubate 2 minutes on ice
- Add 2 ml of LB medium and incubate 60 minutes at 37°C with an agitation at 200 rpm
- Plate 200 µl of BBa_K823012 or BBa_J23101 on a chloramphenicol LB agar plate.
- Incubate plate overnight at 37°C

Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
Miniprep cultures from the 19th August was purified with the NucleoSpin Plasmid kit (Macherey-Nagel). DNA was quantify with the Nanodrop 2000. DNA concentrations and A260/280 were respectively for BBa_E0240 and BBa_I20260: 42 ng/µl and 1.91 ; 13.7 ng/µl and 1.71.
A glycerol stock was prepared for each culture and stored at -80°C.

Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
Preparation of a 1% agarose gel: 1.01 g of Top Vision agarose (Thermo Scientific) + 100 ml of TAE 1X. Microwave 30s by 30s until agarose total dissolution Gel was cooling down until to be lukewarm, one BET drop was added. Gel was loaded with 10µl per sample previously added with 2 µl of loading dye 6X, and 5 µl for ladders. Gel running 45 minutes at 100 mV in TAE 1X buffer.
Purified PCR product aliquot was send to sequencing. The sequencing number were 26DJ68, 69, 70, 71.
Preparation of miniprep culture of BBa_I20260 and BBa_E0240 selected colonies in 5 ml of LB with respectively 5 µl of Kana or chloramphenicol solution stock. Incubation overnight at 37°C and 300 rpm. Aug 19

Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
Sequencing results were received for BBa_J23101 (26DJ62 and 26DJ63) and BBa_K823012(26DJ64 and 26DJ65) PCR products.
26DJ62
TCAGANNAAAAAAATCCTTAGCTNNCGCTAAGGATGANNTCTGGAATTCGCGGCCGCTTCTAGAGTTTACAGCTAGCTCAGTCCTAGGTATTATGCTAGCTACTAGTAG CGGCCGCTGCAGTCCGGCAAAAAAGGGCAAGGTGTCACCACCCTGCCCTTTTTCTTTAAAACCGAAAAGATTACTTCGCGTTATGCAGGCTTCCTCGCTCACTGACTC GCTGCGCTCGGTCGTTCGGCTGCGGCGAGCGGTATCAGCTCACTCAGAG
26DJ63
GAGCGCANCGAGTCAGTGAGCGAGGAAGCCTGCATAACGCGAAGTAATCTTTTCGGTTTTAAAGAAAAAGGGCAGGGTGGTGACACCTTGCCCTTTTTTGCCGGACTG CAGCGGCCGCTACTAGTAGCTAGCATAATACCTAGGACTGAGCTAGCTGTAAACTCTAGAAGCGGCCGCGAATTCCAGAAATCATCCTTAGCGAAAGCTAAGGATTTTT TTTATCTGAAATTCTGCCTCGTGATACGCCTATTTTTATAGGTTAATGTCATGATAATAATGGTTTCTTAGACGCAAGGTGGCAAA
26DJ64
TCAGANNAAAAAAATCCTTAGCTNNCGCTAAGGATGATTTCTGGAATTCGCGGCCGCTTCTAGAGGTTTATAGCTAGCTCATCCTAGGTACAATGCTAGCTACTAG TAGCGGCCGCTGCAGTCCGGCAAAAAAGGGCAAGGTGTCACCACCCTGCCCTTTTTCTTTAAAACCGAAAAGATTACTTCGCGTTATGCAGGCTTCCTCGCTCACTGAC TCGCTGCGCTCGGTCGTTCGGCTGCGGCGAGCGGTATCAGCTCACTCAGG
26DJ65
CNGCGAGTCAGTNAGCGAGGAAGCCTGCANNACGCGAAGTNATCTTTTTCGGTTTTAAAGAAAAAGGGCAGGGTGGTGACACCTNGCCCTTTTTTGCCGGACTGCAGC GGCCGCTACTAGTAGCTAGCATTGTACCTAGGACTGAGCTAGCTATAAACTCTAGAAGCGGCCGCGAATTCCAGAAATCATCCTTAGCGAAAGCTAAGGATTTTTTTTA TCTGAAATTCTGCCTCGTGATACGCCTATTTTTATAGGTTAATGTCATGATAATAATGGTTTCTTAAC
The mapping was perfect for BBa_J23101 for BBa_K823012.
Sequencing results were received for BBa_E0240 (26DJ68 and 26DJ69) and BBa_I20260 (26DJ70 and 26DJ71) PCR products.
26DJ68
GAGCNCAGCGAGTCAGTGAGCGAGGAAGCCTGCATAACGCGAAGTAATCTTTTCGGTTTTAAAGAAAAAGGGCAGGGTGGTGACACCTTGCCCTTTTTTGCCGGACTG CAGCGGCCGCTACTAGTATATAAACGCAGAAAGGCCCACCCGAAGGTGAGCCAGTGTGACTCTAGTAGAGAGCGTTCACCGACAAACAACAGATAAAACGAAAGGCCC AGTCTTTCGACTGAGCCTTTCGTTTTATTTGATGCCTGGCTCTAGTATTATTATTTGTATAGTTCATCCATGCCATGTGTAATCCCAGCAGCTGTTACAAACTCAAGA AGGACCATGTGGTCTCTCTTTTCGTTGGGATCTTTCGAAAGGGCAGATTGTGTGGACAGGTAATGGTTGTCTGGTAAAAGGACAGGGCCATCGCCAATTGGAGTATTT TGTTGATAATGGTCTGCTAGTTGAACGCTTCCATCTTCAATTTGTGTCTAATTTTGAAGTTAACTTTGATTCCATTCTTTTGTTTGTCTGCCATGATGTATACATTGT GTGAGTTATAGTTGTATTCCAATTTGTGTCCAAGAATGTTTCCATCTTCTTTAAAATCAATACCTTTTAACTCGATTCTATTAACAAGGGTATCACCTTCAAACTTGA CTTCAGCACGTGTCTTGTAGTTCCCGTCATCTTTGAAAAATATAGTTCTTTCCTGTACATAACCTTCGGGCATGGCACTCTTGAAAAAGTCATGCTGTTTCATATGAT CTGGGTATCTCGCAAAGCATTGAACACCATAACCGAAAGTAGTGACAAGTGTTGGCCATGGAACAGGTAGTTTTCCAGTAGTGCAAATAAATTTAAGGGTAAGTTTTC CGTATGTTGCATCACCTTCACCCTCTCCACTGACAGAAAATTTGTGCCCATTAACATCACCATCTAATTCAACAAGAATTGGGACAACTCCAGTGAANAGTTCTTCTC CTTTACGCATCTAGTACTTTCCTGTGTGACTCTAGTAGCTAGCATAATACCTAGGACTGAGCTAGCTGTNAACTCTANAAGCGGCCGCGAATTCCAGAAATCATCCTT ANCNNAAGCTAAGGATTTTTTTTATCTGNAATTCTGCCTC
26DJ69
GATTACTATNAAATAGGCGTANNANGAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTAAGGATGATTTCTGGAATTCGCGGCCGCTTCTAGAGTTTACAG CTAGCTCAGTCCTAGGTATTATGCTAGCTACTAGAGTCACACAGGAAAGTACTAGATGCGTAAAGGAGAAGAACTTTTCACTGGAGTTGTCCCAATTCTTGTTGAATT AGATGGTGATGTTAATGGGCACAAATTTTCTGTCAGTGGAGAGGGTGAAGGTGATGCAACATACGGAAAACTTACCCTTAAATTTATTTGCACTACTGGAAAACTACC TGTTCCATGGCCAACACTTGTCACTACTTTCGGTTATGGTGTTCAATGCTTTGCGAGATACCCAGATCATATGAAACAGCATGACTTTTTCAAGAGTGCCATGCCCGA AGGTTATGTACAGGAAAGAACTATATTTTTCAAAGATGACGGGAACTACAAGACACGTGCTGAAGTCAAGTTTGAAGGTGATACCCTTGTTAATAGAATCGAGTTAAA AGGTATTGATTTTAAAGAAGATGGAAACATTCTTGGACACAAATTGGAATACAACTATAACTCACACAATGTATACATCATGGCAGACAAACAAAAGAATGGAATCAA AGTTAACTTCAAAATTAGACACAACATTGAAGATGGAAGCGTTCAACTAGCAGACCATTATCAACAAAATACTCCAATTGGCGATGGCCCTGTCCTTTTACCAGACAA CCATTACCTGTCCACACAATCTGCCCTTTCGAAAGATCCCAACGAANAGAGAGACCACATGGTCCTTCTTGAGTTTGTAACAGCTGCTGGGATTACACATGGCATGGA TGAACTATACAAATAATAATACTAGAGCCAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCGGTGAACGCTCTCT ACTAGAGTCACACTGGCTCACCTTCGGGTGGGCCTTTCTGCGTTTATATACTAGTAGCGGCCGCTGCAGTCCGGCNAAAAAGGGCAAGGTGTCACCACCNTGCCCTTT TTCTTTAAAACCGNAAA
26DJ70
AGCGAGTCAGTGAGCGAGGAAGCCTGCATAACGCGAAGTAATCTTTTCGGTTTTAAAGAAAAAGGGCAGGGTGGTGACACCTTGCCCTTTTTTGCCGGACTGCAGCGG CCGCTACTAGTATATAAACGCAGAAAGGCCCACCCGAAGGTGAGCCAGTGTGACTCTAGTAGAGAGCGTTCACCGACAAACAACAGATAAAACGAAAGGCCCAGTCTT TCGACTGAGCCTTTCGTTTTATTTGATGCCTGGCTCTAGTATTATTATTTGTATAGTTCATCCATGCCATGTGTAATCCCAGCAGCTGTTACAAACTCAAGAAGGACC ATGTGGTCTCTCTTTTCGTTGGGATCTTTCGAAAGGGCAGATTGTGTGGACAGGTAATGGTTGTCTGGTAAAAGGACAGGGCCATCGCCAATTGGAGTATTTTGTTGA TAATGGTCTGCTAGTTGAACGCTTCCATCTTCAATGTTGTGTCTAATTTTGAAGTTAACTTTGATTCCATTCTTTTGTTTGTCTGCCATGATGTATACATTGTGTGAG TTATAGTTGTATTCCAATTTGTGTCCAAGAATGTTTCCATCTTCTTTAAAATCAATACCTTTTAACTCGATTCTATTAACAAGGGTATCACCTTCAAACTTGACTTCA GCACGTGTCTTGTAGTTCCCGTCATCTTTGAAAAATATAGTTCTTTCCTGTACATAACCTTCGGGCATGGCACTCTTGAAAAAGTCATGCTGTTTCATATGATCTGGG TATCTCGCAAAGCATTGAACACCATAACCGAAAGTAGTGACAAGTGTTGGCCATGGAACAGGTAGTTTTCCAGTAGTGCAAATAAATTTAAGGGTAAGTTTTCCGTAT GTTGCATCACCTTCACCCTCTCCACTGACAGAAAATTTGTGCCCATTAACATCACCATCTAATTCAACAAGAATTGGGACAACTCCAGTGAAAAGTTCTTCTCCTTTA CGCATCTAGTACTTTCCTGTGTGACTCTAGAAGCGGCCGCGAATTCCAGAAATCATCCTTAGCGATGAATTCCAGAAATCATCCTTANCGAAAGCTNAGGATTTTTTT TATCTGNAATTCTGNCTCG
26DJ71
GATTAAACTATNAATAGGCGTATCACGAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTAAGGATGATTTCTGGAATTCATCGCTAAGGATGATTTCTGGA ATTCGCGGCCGCTTCTAGAGTCACACAGGAAAGTACTAGATGCGTAAAGGAGAAGAACTTTTCACTGGAGTTGTCCCAATTCTTGTTGAATTAGATGGTGATGTTAAT GGGCACAAATTTTCTGTCAGTGGAGAGGGTGAAGGTGATGCAACATACGGAAAACTTACCCTTAAATTTATTTGCACTACTGGAAAACTACCTGTTCCATGGCCAACA CTTGTCACTACTTTCGGTTATGGTGTTCAATGCTTTGCGAGATACCCAGATCATATGAAACAGCATGACTTTTTCAAGAGTGCCATGCCCGAAGGTTATGTACAGGAA AGAACTATATTTTTCAAAGATGACGGGAACTACAAGACACGTGCTGAAGTCAAGTTTGAAGGTGATACCCTTGTTAATAGAATCGAGTTAAAAGGTATTGATTTTAAA GAAGATGGAAACATTCTTGGACACAAATTGGAATACAACTATAACTCACACAATGTATACATCATGGCAGACAAACAAAAGAATGGAATCAAAGTTAACTTCAAAATT AGACACAACATTGAAGATGGAAGCGTTCAACTAGCAGACCATTATCAACAAAATACTCCAATTGGCGATGGCCCTGTCCTTTTACCAGACAACCATTACCTGTCCACA CAATCTGCCCTTTCGAAAGATCCCAACGAANAGAGAGACCACATGGTCCTTCTTGAGTTTGTAACAGCTGCTGGGATTACACATGGCATGGATGAACTATACAAATAA TAATACTAGAGCCAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCGGTGAACGCTCTCTACTAGAGTCACACTGG CTCACCTTCGGGTGGGCCTTTCTGCGTTTATATACTAGTAGCGGCCGCTGCAGTCCGGCAAANAAGGGCAAGGTGTCACCACCNTGCCCTTTTTCTTTAAAACCGNAA AGATTACTTCNNNGTTATGCAGGCTTCCTCGCTCACTGACTCGCTGCGCTCGGTCGTTCGGCT
The mapping were correct for BBa_E0240 and BBa_I20260.
Colony PCRs were performed on 8 colonies for BBa_E0240 and BBa_I20260, form plates from the transformation of 16th August. 49 µl of mix were distributed in each PCR tubes. The One Taq PCR program was applied. Aug 18

Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
Colonies had grown on plates as followed:
- LB agar: more than 1000 colonies for the two biobricks
- LB agar kanamycin: 10 colonies for BBa_E0240 and >40 colonies for BBa_I20260
- LB agar chloramphenicol: >50 colonies for BBa_E0240 and 3 colonies for BBa_I20260
Plate storage at 4°C.
Aug 17
Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
Nothing had grown during the night on both plates. Taking into consideration different hypothesis, the transformation was repeated with a change on step 6 and 7. At step 6, only 1 ml of LB medium was added. To plate, 200 µl of BBa_E0240 and BBa_I20260 were plated on 3 plates: LB agar, LB agar kanamycin and LB agar chloramphenicol.
Aug 16
Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
BBa_E0240 and BBa_I20260 are respectively into a PSB1C3 and a PSB1K3 vector. In order to select colonies, LB complemented with kanamycin or chloramphenicol are necessary. 200 ml of LB agar medium was prepared with 7 g of LB Agar powder(Sigma) into 200 ml of miliQ water. After complete dissolution, the solution was sterilized in the Tuttmauer 2540ML apparatus, STE 20 minutes at 121°C and EXT+DRY 15 minutes. 50 µl of kanamycin stock solution (25 mg/L) was added to 50 ml of LB agar to pour 2 plates. 150 µl of chloramphenicol stock solution was added in the 150 LB agar remaining ml to pour 6 plates.
The transformation was performed into DH5 alpha ''E. coli'', as followed:
- Remove E. coli competent tubes from -80°C and keep it on ice
- Add 1 µl of template (here solubilized plasmids from the registry distribution kit) and mix gently
- Incubate 10 minutes on ice
- Perform an heat shock 30 seconds at 42°C
- Incubate 2 minutes on ice
- Add 3 ml of LB medium and incubate 60 minutes at 37°C with an agitation at 200 rpm
- Plate 200 µl of BBa_E0240 on a chloramphenicol LB agar plate and BBa_I20260 on a Knamycin LB agar plate
- Incubate plate overnight at 37°C

Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
To amplify BBa_I20260 and BBa_0240, a PCR was perform with the mix described Table 1. The program IGEM Q5 PCR was applied, see Table 2. PCR products were cleaned with the GeneJET purification kit (Thermo Scientific)followed by DNA quantification with the NanoDrop 2000 (Thermo Scientific). Purified samples of I20260 and E1010 had respectively following concentration 30.9 ng/µl and 43.3 ng/µl.
Preparation of a 1% agarose gel: 0.52 g of Top Vision agarose (Thermo Scientific) + 50 ml of TAE 1X. Microwave 30s by 30s until agarose total dissolution Gel was cooling down until to be lukewarm, one BET drop was added. Gel was loaded with 20 µl of BBa_E0240 PCR product and BBa_I20260 purified PCR product added with 4 µl of loading dye, at 10µl of the mix per well.
Gel running 45 minutes at 100 mV in TAE 1X buffer.
3 pieces of gel were sampled in four tubes to perform a DNA purification from gel. The DNA purification was made with the GeneJET purification kit (Thermo Scientific).
A verification electrophoresis was performed on a 1% agarose gel.
The major part of the DNA was lost during the purification because the amount of was to weak to be purified from gel. So we decide do transform ''E. coli'' to isolate a colony containing the desired plasmid for BBa_E0240 and BBa_I20260.
Aug 14
Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
PCR products were cleaned with the GeneJET purification kit (Thermo Scientific)followed by DNA quantification with the NanoDrop 2000 (Thermo Scientific).
Purified PCR product of BBa_J61002/J23101 and BBa_J61002/K823012 concentration were respectively: 53.9 ng/µl and 54.1 ng/µl.
Preparation of a 1% agarose gel: 0.56 g of Top Vision agarose (Thermo Scientific) + 50 ml of TAE 1X. Microwave 30s by 30s until agarose total dissolution Gel was cooling down until to be lukewarm, one BET drop was added. Gel was loaded with 10µl per sample previously added with 2 µl of loading dye 6X, and 5 µl for ladders. Gel running 45 minutes at 100 mV in TAE 1X buffer.
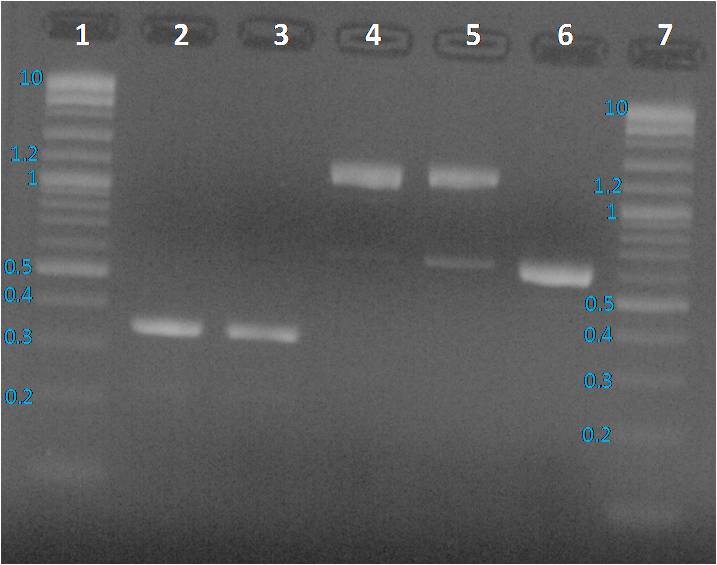
Profiles were same as before PCR clean up. To sequence the amplified parts, we decided to a purification of fragments from the gel as presented below.
Preparation of a 1% agarose gel: 0.56 g of Top Vision agarose (Thermo Scientific) + 50 ml of TAE 1X. Microwave 30s by 30s until agarose total dissolution. Gel was cooling down until to be lukewarm, one BET drop was added. Gel was loaded with 30 µl of BBa_E0240 purified PCR product and BBa_I20260 purified PCR product added with 6 µl of loading dye, at 10µl of the mix per well. Gel running 45 minutes at 100 mV in TAE 1X buffer.
Four pieces of gel were sampled in four tubes to perform a DNA purification from gel.
Aug 13

Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
The 4 needed parts are: BBa_J23101, BBa_K823012, BBa_E0240 and BBa_I20260. Corresponding wells were located on 2014 Distribution kit plates and resuspended with 10 µL steril water. That permits to obtain a DNA concentration around 0.2 ng/µl (according to the registry. Solutions were transferred into 1 ml eppendorf tubes and stored at -20°C.
To amplify fragments, a PCR was performed on the 4 constructions, with the mix described on table 1.
Distribution of 49 µl of mix per PCR tube. Application of program IGEM Q5 PCR.
Preparation of a 1% agarose gel: 0.56 g of Top Vision agarose (Thermo Scientific) + 50 ml of TAE 1X.Microwave 30s by 30s until agarose total dissolution. Gel was cooling down until to be lukewarm, one BET drop was added. Gel was loaded with 10µl per sample previously added with 2 µl of loading dye 6X, and 5 µl for ladders. Gel running 45 minutes at 100 mV in TAE 1X buffer.
We expected to obtain one band per PCR sample corresponding to the interesting amplified fragment. For Lane 2, 3 and 6 it was ok. We had the expected profile. By contrast 2 bands were visible, one at the expected size (around 1200 bp) and another around 600 bp. We decided to perform a purification on gel of each bands.
Aug 12
 "
"










