Team:KIT-Kyoto/Notebook/Labnote
From 2014.igem.org
| Line 3,875: | Line 3,875: | ||
<br> | <br> | ||
</p> | </p> | ||
| + | |||
<h3>August 20, 2014</h3> | <h3>August 20, 2014</h3> | ||
| Line 3,959: | Line 3,960: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>Mu-ta-CuMTS3</td> | <td>Mu-ta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Sal</em>1</td> |
<td>Fast Di-gest buffer</td> | <td>Fast Di-gest buffer</td> | ||
</tr> | </tr> | ||
| Line 3,966: | Line 3,967: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>Mu-ta-CuMTS3</td> | <td>Mu-ta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Sal</em>1</td> |
<td>Fast Di-gest buffer</td> | <td>Fast Di-gest buffer</td> | ||
</tr> | </tr> | ||
| Line 3,973: | Line 3,974: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>Mu-ta-CuMTS3</td> | <td>Mu-ta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Sal</em>1</td> |
<td>Fast Di-gest buffer</td> | <td>Fast Di-gest buffer</td> | ||
</tr> | </tr> | ||
| Line 3,980: | Line 3,981: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>Mu-ta-CuMTS3</td> | <td>Mu-ta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Sal</em>1</td> |
<td>Fast Di-gest buffer</td> | <td>Fast Di-gest buffer</td> | ||
</tr> | </tr> | ||
| Line 4,017: | Line 4,018: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Sal</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 4,025: | Line 4,026: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Sal</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 4,033: | Line 4,034: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Sal</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 4,041: | Line 4,042: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Sal</em>1</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 4,065: | Line 4,066: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Fast Di-gest buffer</td> | <td>Fast Di-gest buffer</td> | ||
</tr> | </tr> | ||
| Line 4,072: | Line 4,073: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>Mu-ta-CuMTS3</td> | <td>Mu-ta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Fast Di-gest buffer</td> | <td>Fast Di-gest buffer</td> | ||
</tr> | </tr> | ||
| Line 4,106: | Line 4,107: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>CuMTS3</td> | <td>CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 4,113: | Line 4,114: | ||
<td>pGEX6P-2</td> | <td>pGEX6P-2</td> | ||
<td>Mu-ta-CuMTS3</td> | <td>Mu-ta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 4,290: | Line 4,291: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>BBa_J04450</td> | <td>BBa_J04450</td> | ||
| - | <td> | + | <td><em>Eco</em>R1, <em>Pst</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 4,302: | Line 4,303: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>BBa_J04450</td> | <td>BBa_J04450</td> | ||
| - | <td> | + | <td></td> |
<td></td> | <td></td> | ||
</tr> | </tr> | ||
| Line 4,339: | Line 4,340: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>BBa_J04450</td> | <td>BBa_J04450</td> | ||
| - | <td> | + | <td><em>Eco</em>R1, <em>Pst</em>1/td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 4,355: | Line 4,356: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>BBa_J04450</td> | <td>BBa_J04450</td> | ||
| - | <td> | + | <td>/td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 4,417: | Line 4,418: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td></td> | <td></td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td><em>Xba</em>1, <em>Spe</em>1</td> |
<td>Fast Digest</td> | <td>Fast Digest</td> | ||
</tr> | </tr> | ||
| Line 4,425: | Line 4,426: | ||
<td>muta-CuMTS3</td> | <td>muta-CuMTS3</td> | ||
<td>terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1</td> | <td>terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1</td> | ||
| - | <td> | + | <td><em>Xba</em>1, <em>Spe</em>1</td> |
<td>Fast Digest</td> | <td>Fast Digest</td> | ||
</tr> | </tr> | ||
| Line 4,469: | Line 4,470: | ||
<td>muta-CuMTS3</td> | <td>muta-CuMTS3</td> | ||
<td>terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1</td> | <td>terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1</td> | ||
| - | <td> | + | <td><em>Spe</em>1, <em>Xba</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 4,884: | Line 4,885: | ||
<td></td> | <td></td> | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td></td> |
| - | <td> | + | <td></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 4,893: | Line 4,894: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 4,901: | Line 4,902: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 4,909: | Line 4,910: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 4,917: | Line 4,918: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 4,924: | Line 4,925: | ||
<td>DH5α (セ)</td> | <td>DH5α (セ)</td> | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td><em>Eco</em>R1, <em>Pst</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 4,933: | Line 4,934: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1, <em>Pst</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 4,941: | Line 4,942: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1, <em>Pst</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 4,949: | Line 4,950: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1, <em>Pst</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 4,957: | Line 4,958: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1, <em>Pst</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 4,964: | Line 4,965: | ||
<td></td> | <td></td> | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td><em>Xba</em>1</td> |
<td>FD buffer</td> | <td>FD buffer</td> | ||
</tr> | </tr> | ||
| Line 4,973: | Line 4,974: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Xba</em>1</td> |
<td>FD buffer</td> | <td>FD buffer</td> | ||
</tr> | </tr> | ||
| Line 4,981: | Line 4,982: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Xba</em>1</td> |
<td>FD buffer</td> | <td>FD buffer</td> | ||
</tr> | </tr> | ||
| Line 4,989: | Line 4,990: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Xba</em>1</td> |
<td>FD buffer</td> | <td>FD buffer</td> | ||
</tr> | </tr> | ||
| Line 4,997: | Line 4,998: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Xba</em>1</td> |
<td>FD buffer</td> | <td>FD buffer</td> | ||
</tr> | </tr> | ||
| Line 5,022: | Line 5,023: | ||
<td></td> | <td></td> | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 5,031: | Line 5,032: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 5,039: | Line 5,040: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 5,047: | Line 5,048: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 5,055: | Line 5,056: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 5,071: | Line 5,072: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1, <em>Pst</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 5,079: | Line 5,080: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1, <em>Pst</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 5,087: | Line 5,088: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1, <em>Pst</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 5,095: | Line 5,096: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1, <em>Pst</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 5,103: | Line 5,104: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1, <em>Pst</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 5,118: | Line 5,119: | ||
<td></td> | <td></td> | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td><em>Xba</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 5,127: | Line 5,128: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Xba</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 5,135: | Line 5,136: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Xba</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 5,143: | Line 5,144: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Xba</em>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 5,151: | Line 5,152: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Xba</em>1</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 5,240: | Line 5,241: | ||
<tr> | <tr> | ||
<th colspan="5">Sample</th> | <th colspan="5">Sample</th> | ||
| - | <th | + | <th>Enzyme</th> |
| - | <th | + | <th>Buffer</th> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 5,264: | Line 5,265: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>BBa_J04450</td> | <td>BBa_J04450</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 5,273: | Line 5,274: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>BBa_J04450</td> | <td>BBa_J04450</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td><em>Pst</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 5,282: | Line 5,283: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>BBa_J04450</td> | <td>BBa_J04450</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td><em>Eco</em>R1, <em>Pst</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 5,291: | Line 5,292: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>BBa_J04450</td> | <td>BBa_J04450</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td><em>Spe</em>1</td> |
<td>FD buffer</td> | <td>FD buffer</td> | ||
</tr> | </tr> | ||
| Line 5,300: | Line 5,301: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>BBa_J04450</td> | <td>BBa_J04450</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td><em>Xba</em>1</td> |
<td>FD buffer</td> | <td>FD buffer</td> | ||
</tr> | </tr> | ||
| Line 5,309: | Line 5,310: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>BBa_J04450</td> | <td>BBa_J04450</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xba</em>1</td> |
<td>FD buffer</td> | <td>FD buffer</td> | ||
</tr> | </tr> | ||
| Line 5,318: | Line 5,319: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>BBa_J04450</td> | <td>BBa_J04450</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td></td> |
<td></td> | <td></td> | ||
</tr> | </tr> | ||
| Line 5,336: | Line 5,337: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Mu-ta-CuMTS3</td> | <td>Mu-ta-CuMTS3</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td><em>Eco</em>R1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 5,345: | Line 5,346: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Mu-ta-CuMTS3</td> | <td>Mu-ta-CuMTS3</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td><em>Pst</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 5,354: | Line 5,355: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Mu-ta-CuMTS3</td> | <td>Mu-ta-CuMTS3</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td><em>Eco</em>R1, <em>Pst</em>1</td> |
<td>Buffer H</td> | <td>Buffer H</td> | ||
</tr> | </tr> | ||
| Line 5,363: | Line 5,364: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Mu-ta-CuMTS3</td> | <td>Mu-ta-CuMTS3</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td><em>Spe</em>1</td> |
<td>FD buffer</td> | <td>FD buffer</td> | ||
</tr> | </tr> | ||
| Line 5,372: | Line 5,373: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Mu-ta-CuMTS3</td> | <td>Mu-ta-CuMTS3</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td><em>Xba</em>1</td> |
<td>FD buffer</td> | <td>FD buffer</td> | ||
</tr> | </tr> | ||
| Line 5,381: | Line 5,382: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Mu-ta-CuMTS3</td> | <td>Mu-ta-CuMTS3</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td><em>Spe</em>1, <em>Xba</em>1</td> |
<td>FD buffer</td> | <td>FD buffer</td> | ||
</tr> | </tr> | ||
| Line 5,390: | Line 5,391: | ||
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Mu-ta-CuMTS3</td> | <td>Mu-ta-CuMTS3</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td></td> |
<td></td> | <td></td> | ||
</tr> | </tr> | ||
| Line 5,401: | Line 5,402: | ||
<table class="materials"> | <table class="materials"> | ||
<tr> | <tr> | ||
| - | <th | + | <th>Lane</th> |
| - | <th | + | <th rowspan="4">Sample</th> |
| - | <th | + | <th>Enzyme</th> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 5,410: | Line 5,411: | ||
<th>Vector</th> | <th>Vector</th> | ||
<th>Insert</th> | <th>Insert</th> | ||
| - | |||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>1</td> | <td>1</td> | ||
| - | |||
| - | |||
<td></td> | <td></td> | ||
<td></td> | <td></td> | ||
| + | <td>pSB1C3</td> | ||
<td></td> | <td></td> | ||
<td></td> | <td></td> | ||
| Line 5,424: | Line 5,423: | ||
<td>2</td> | <td>2</td> | ||
<td></td> | <td></td> | ||
| - | <td></td> | + | <td>DH5α (セ)</td> |
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
| - | <td> | + | <td>Muta-CuMTS3</td> |
| - | <td> | + | <td></td> |
| - | + | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>3</td> | <td>3</td> | ||
<td></td> | <td></td> | ||
| - | <td></td> | + | <td>DH5α (ソ)</td> |
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
| - | <td> | + | <td>Muta-CuMTS3</td> |
| - | <td> | + | <td></td> |
| - | + | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>4</td> | <td>4</td> | ||
<td></td> | <td></td> | ||
| - | <td></td> | + | <td>DH5α (F)</td> |
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
| - | <td> | + | <td>Muta-CuMTS3</td> |
| - | <td> | + | <td></td> |
| - | + | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>5</td> | <td>5</td> | ||
<td></td> | <td></td> | ||
| - | <td></td> | + | <td>DH5α (N)</td> |
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
| - | <td> | + | <td>Muta-CuMTS3</td> |
| - | <td> | + | <td></td> |
| - | + | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>6</td> | <td>6</td> | ||
| + | <td>1kb Ladder</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
<td></td> | <td></td> | ||
<td></td> | <td></td> | ||
| - | |||
| - | |||
| - | |||
| - | |||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>7</td> | <td>7</td> | ||
<td></td> | <td></td> | ||
| - | <td></td> | + | <td>DH5α (セ)</td> |
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
| - | <td> | + | <td>Muta-CuMTS3</td> |
| - | <td> | + | <td><em>Eco</em>R1, <em>Pst</em>1</td> |
| - | < | + | |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>8</td> | <td>8</td> | ||
<td></td> | <td></td> | ||
| - | <td></td> | + | <td>DH5α (ソ)</td> |
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
| - | <td> | + | <td>Muta-CuMTS3</td> |
| - | <td> | + | <td><em>Eco</em>R1, <em>Pst</em>1</td> |
| - | < | + | |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>9</td> | <td>9</td> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<td></td> | <td></td> | ||
| + | <td>DH5α (F)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>Muta-CuMTS3</td> | ||
| + | <td><em>Eco</em>R1, <em>Pst</em>1</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>10</td> | <td>10</td> | ||
<td></td> | <td></td> | ||
| - | <td> | + | <td>DH5α (N)</td> |
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1, <em>Pst</em>1</td> |
| - | < | + | |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>11</td> | <td>11</td> | ||
<td></td> | <td></td> | ||
| - | <td> | + | <td></td> |
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Eco</em>R1, <em>Pst</em>1</td> |
| - | < | + | |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>12</td> | <td>12</td> | ||
| + | <td>1kb Ladder</td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
<td></td> | <td></td> | ||
<td></td> | <td></td> | ||
| - | |||
| - | |||
| - | |||
| - | |||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>13</td> | <td>13</td> | ||
<td></td> | <td></td> | ||
| - | <td> | + | <td></td> |
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
| - | <td> | + | <td></td> |
| - | <td> | + | <td><em>Xba</em>1</td> |
| - | < | + | |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>14</td> | <td>14</td> | ||
<td></td> | <td></td> | ||
| - | <td> | + | <td>DH5α (セ)</td> |
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Xba</em>1</td> |
| - | < | + | |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>15</td> | <td>15</td> | ||
<td></td> | <td></td> | ||
| - | <td> | + | <td>DH5α (ソ)</td> |
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Xba</em>1</td> |
| - | < | + | |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>16</td> | <td>16</td> | ||
<td></td> | <td></td> | ||
| - | <td>F</td> | + | <td>DH5α (F)</td> |
<td>pSB1C3</td> | <td>pSB1C3</td> | ||
<td>Muta-CuMTS3</td> | <td>Muta-CuMTS3</td> | ||
| - | <td> | + | <td><em>Xba</em>1</td> |
| - | <td> | + | </tr> |
| + | <tr> | ||
| + | <td>17</td> | ||
| + | <td></td> | ||
| + | <td>DH5α (N)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>Muta-CuMTS3</td> | ||
| + | <td><em>Xba</em>1</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| Line 5,560: | Line 5,552: | ||
<br> | <br> | ||
</tr> | </tr> | ||
| + | <br> | ||
| + | Sample F was selected as a submission plasmid. | ||
| + | <br> | ||
| + | <strong>Main Culture</strong> | ||
| + | <br> | ||
| + | <div class="m_table"> | ||
| + | <table class="materials"> | ||
| + | <tr> | ||
| + | <th colspan="4">Sample</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Host</th> | ||
| + | <th>Vector</th> | ||
| + | <th>Insert</th> | ||
| + | <th>Primer</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α(F)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>mu-ta-CuMTS3</td> | ||
| + | <td>----------</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α(F)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>mu-ta-CuMTS3</td> | ||
| + | <td>----------</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DH5α(F)</td> | ||
| + | <td>pSB1C3</td> | ||
| + | <td>mu-ta-CuMTS3</td> | ||
| + | <td>----------</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | Cultivated these samples in 3mL of LB medium with ChlPhe at 30°C overnight. | ||
| + | <br> | ||
</p> | </p> | ||
| + | |||
| + | <h3> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | <br>a | ||
| + | <br>a | ||
| + | <br>a | ||
| + | <br>a | ||
| + | <br>a | ||
| + | <br> | ||
| + | <br><br> | ||
| + | <br> | ||
| + | <br>a | ||
| + | <br>a | ||
| + | <br>a | ||
| + | <br>a | ||
| + | <br>a | ||
| + | <br> | ||
| + | <br><br> | ||
| + | <br> | ||
| + | <br>a | ||
| + | <br>a | ||
| + | <br>a | ||
| + | <br>a | ||
| + | <br>a | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br></p> | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 5,569: | Line 5,632: | ||
<div class="ac"> | <div class="ac"> | ||
<p class="sentence"> | <p class="sentence"> | ||
| - | |||
<h3>October 1, 2014</h3> | <h3>October 1, 2014</h3> | ||
Revision as of 06:02, 16 October 2014


LabNote
June
June 10, 2014
OKAMOTO
Miniprep
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| BL21(DE3) | pGEX6P-2 | CuMTSE1 |
| BL21(DE3) | pGEX6P-2 | CuMTSE2 |
| BL21(DE3) | pGEX6P-2 | CuMTS3 |
| BL21(DE3) | pGEX6P-2 | CuMTS62 |
Restriction Digest
| Host | Sample | Enzyme | Buffer | |
|---|---|---|---|---|
| Vector | Insert | |||
| BL21(DE3) | pGEX6P-2 | CuMTSE1 | EcoR1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTSE2 | EcoR1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTS3 | EcoR1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTS62 | EcoR1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTSE1 | Sal1, Not1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTSE2 | Sal1, Not1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTS3 | Sal1, Not1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTS62 | Sal1, Not1 | Buffer H |
June 11, 2014
KOBAYASHI
AGE
| Lane | Sample | Enzyme | ||
|---|---|---|---|---|
| Marker | Vector | Insert | ||
| 1 | λDNA-HindⅢ | |||
| 2 | pGEX6P-2 | CuMTSE1 | N.D | |
| 3 | pGEX6P-2 | CuMTSE1 | EcoR1 | |
| 4 | pGEX6P-2 | CuMTSE1 | Sal1, Not1 | |
| 5 | pGEX6P-2 | CuMTSE2 | N.D | |
| 6 | pGEX6P-2 | CuMTSE2 | EcoR1 | |
| 7 | pGEX6P-2 | CuMTSE2 | Sal1, Not1 | |
| 8 | pGEX6P-2 | CuMTS3 | N.D | |
| 9 | pGEX6P-2 | CuMTS3 | EcoR1 | |
| 10 | pGEX6P-2 | CuMTS3 | Sal1, Not1 | |
| 11 | pGEX6P-2 | CuMTS62 | N.D | |
| 12 | pGEX6P-2 | CuMTS62 | EcoR1 | |
| 13 | pGEX6P-2 | CuMTS62 | Sal1, Not1 | |

June 12, 2014
KOBAYASHI
Transformation
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS3 |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS62 |
June 13, 2014
KOBAYASHI
Colony Isolation
Select 6 colonies per plate from 4 plates from June 12.
| Host | Sample | Section | Enzyme | |
|---|---|---|---|---|
| Vector | Insert | |||
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | 1-a, 1-b | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | 2-a, 2-b | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS3 | 3-a, 3-b | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS62 | 6-a, 6-b | |
June 14, 2014
ONISHI
Miniprep
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| 1-a | pGEX6P-2 | CuMTSE1 |
| 1-b | pGEX6P-2 | CuMTSE1 |
| 2-a | pGEX6P-2 | CuMTSE2 |
| 2-b | pGEX6P-2 | CuMTSE2 |
| 3-a | pGEX6P-2 | CuMTS3 |
| 3-b | pGEX6P-2 | CuMTS3 |
| 6-a | pGEX6P-2 | CuMTS62 |
| 6-b | pGEX6P-2 | CuMTS62 |
Miniprep
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| BL21(DE3) | pGEX6P-2 | CuMTSE1 |
| BL21(DE3) | pGEX6P-2 | CuMTSE2 |
| BL21(DE3) | pGEX6P-2 | CuMTS3 |
| BL21(DE3) | pGEX6P-2 | CuMTS62 |
Restriction Digest
| Host | Sample | Enzyme | Buffer | |
|---|---|---|---|---|
| Vector | Insert | |||
| 1-a | pGEX6P-2 | CuMTSE1 | EcoR1 | Buffer H |
| 1-b | pGEX6P-2 | CuMTSE1 | EcoR1 | Buffer H |
| 2-a | pGEX6P-2 | CuMTSE2 | EcoR1 | Buffer H |
| 2-b | pGEX6P-2 | CuMTSE2 | EcoR1 | Buffer H |
| 3-a | pGEX6P-2 | CuMTS3 | EcoR1 | Buffer H |
| 3-b | pGEX6P-2 | CuMTS3 | EcoR1 | Buffer H |
| 6-a | pGEX6P-2 | CuMTS62 | EcoR1 | Buffer H |
| 6-b | pGEX6P-2 | CuMTS62 | EcoR1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTSE1 | EcoR1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTSE2 | EcoR1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTS3 | EcoR1 | Buffer H |
| BL21(DE3) | pGEX6P-2 | CuMTS62 | EcoR1 | Buffer H |
AGE
| Lane | Sample | Insert | Enzyme | Buffer | ||
|---|---|---|---|---|---|---|
| Marker | Host | Vector | ||||
| 1 | 1kb Ladder | |||||
| 2 | λDNA-HindⅢ | |||||
| 3 | 1-a | pGEX6P-2 | CuMTSE1 | EcoR1 | Buffer H | |
| 4 | 1-b | pGEX6P-2 | CuMTSE1 | EcoR1 | Buffer H | |
| 5 | 2-a | pGEX6P-2 | CuMTSE2 | EcoR1 | Buffer H | |
| 6 | 2-b | pGEX6P-2 | CuMTSE2 | EcoR1 | Buffer H | |
| 7 | 3-a | pGEX6P-2 | CuMTS3 | EcoR1 | Buffer H | |
| 8 | 3-b | pGEX6P-2 | CuMTS3 | EcoR1 | Buffer H | 9 | 6-a | pGEX6P-2 | CuMTS62 | EcoR1 | Buffer H |
| 10 | 6-b | pGEX6P-2 | CuMTS62 | EcoR1 | Buffer H | |
| 11 | BL21(DE3) | pGEX6P-2 | CuMTSE1 | EcoR1 | Buffer H | |
| 12 | BL21(DE3) | pGEX6P-2 | CuMTSE2 | EcoR1 | Buffer H | |
| 13 | BL21(DE3) | pGEX6P-2 | CuMTS3 | EcoR1 | Buffer H | |
| 14 | BL21(DE3) | pGEX6P-2 | CuMTS62 | EcoR1 | Buffer H | |
Applied λDNA-HindⅢ to compare and contrast AGE of July 14 and June 11.

June 16, 2014
YAMAJI
Preculture
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS3 |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS62 |
| BL21(DE3)pLysS | pGEX6P-2 | |
Main Culture
| Host | Sample | IPTG | |
|---|---|---|---|
| Vector | Insert | ||
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS3 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS62 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | Induced | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Non-induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Non-induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS3 | Non-induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTS62 | Non-induced |
| BL21(DE3)pLysS | pGEX6P-2 | Non-induced | |
June 18, 2014
YAMAJI
Preculture
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 |
Main Culture
| Host | Sample | IPTG | |
|---|---|---|---|
| Vector | Insert | ||
| BL21(DE3)pLysS | pGEX6P-2 | Induced | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | Non-induced | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Non-induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Non-induced |
Protein Extraction (E.coli)
| Host | Sample | IPTG | |
|---|---|---|---|
| Vector | Insert | ||
| BL21(DE3)pLysS | pGEX6P-2 | Induced | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | Non-induced | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Non-induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Non-induced |
Preparations for SDS-PAGE
Add SDS sample buffer to protein extractions in a flask, incubate at 37℃ for 15 minutes and keep it at 4℃.

June 19, 2014
KITOMI
SDS-PAGE
| Host | Sample | IPTG | |
|---|---|---|---|
| Vector | Insert | ||
| BL21(DE3)pLysS | pGEX6P-2 | Induced | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | Non-induced | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Non-induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Non-induced |
June 21, 2014
SOGA
Miniprep
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 |
PCR
| Vector | Sample | DNA Polymerase | |
|---|---|---|---|
| Insert | Primer | ||
| pGEX6P-2 | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | KOD-FX-NEO |
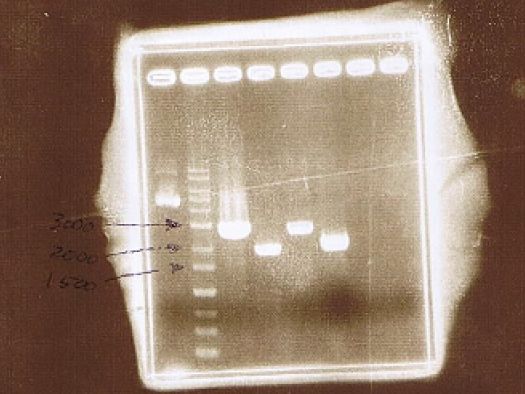
AGE
| Lane | Sample | Insert | Primer | ||
|---|---|---|---|---|---|
| Marker | Host | Vector | |||
| 1 | 1kb Ladder | ||||
| 2 | BY4247 | p427-TEF | |||
| 3 | pGEX6P-2 | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | ||
| 4 | pGEX6P-2 | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | ||
| 5 | pGEX6P-2 | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | ||
| 6 | pGEX6P-2 | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | ||
June 24, 2014
SOGA
Preculture
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| BL21(DE3)pLysS | ||
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 |
June 25, 2014
KOBAYASHI
Cultivated following samples in Big Scale at 30℃ overnight in shaken culture.
| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| BL21(DE3)pLysS | ||
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 |
DNA Refinement1
| Sample | |
|---|---|
| DNA | Primer |
| CuMTSE1 | pGEX6P-2F1, pGEX6P-2R |
| CuMTSE1 | pGEX6P-2F2, pGEX6P-2R |
Restriction Digest
| Vector | Sample | Enzyme | Buffer | |
|---|---|---|---|---|
| DNA | Primer | |||
| CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Buffer H | |
| CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Spe1, Xho1 | Buffer H | |
| p427-TEF | Spe1, Xho1 | Buffer H | ||
DNA Refinement2
| Vector | Sample | Enzyme | |
|---|---|---|---|
| DNA | Primer | ||
| CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | |
| CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Spe1, Xho1 | |
| p427-TEF | Spe1, Xho1 | ||
Main Culture
| Host | Sample | IPTG | |
|---|---|---|---|
| Vector | Insert | ||
| BL21(DE3)pLysS | pGEX6P-2 | Induced | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Induced |
| BL21(DE3)pLysS | pGEX6P-2 | Non-induced | |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 | Non-induced |
| BL21(DE3)pLysS | pGEX6P-2 | CuMTSE2 | Non-induced |

Ligation
| Sample | ||
|---|---|---|
| Vector | DNA | Primer |
| p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R |
| p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R |
| p427-TEF | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R |
| p427-TEF | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R |
Transformation
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| DH5α | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R |
| DH5α | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R |
| DH5α | p427-TEF | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R |
| DH5α | p427-TEF | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R |
June 26, 2014
SAWANO
Colony Isolation
Isolate each colony from the plates which were inoculated on June 25.
| Host | Sample | Section | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| DH5α | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | 1~4 |
| DH5α | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | a~h |
| DH5α | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | ア~タ |
June 27, 2014
SHIMADA
Colony Sweep
| Lane | Sample | ||
|---|---|---|---|
| Host | Vector | Insert | |
| 1 | BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 |
| 2 | 1 | ||
| 3 | 2 | ||
| 4 | 3 | ||
| 5 | 4 | ||
| 6 | a | ||
| 7 | b | ||
| 8 | c | ||
| 9 | d | ||
| 10 | e | ||
| 11 | f | ||
| 12 | g | ||
| 13 | h | ||

Colony Sweep
| Lane | Sample | ||
|---|---|---|---|
| Host | Vector | Insert | |
| 1 | BL21(DE3)pLysS | pGEX6P-2 | CuMTSE1 |
| 2 | ア | ||
| 3 | イ | ||
| 4 | ウ | ||
| 5 | エ | ||
| 6 | オ | ||
| 7 | カ | ||
| 8 | キ | ||
| 9 | ク | ||
| 10 | ケ | ||
| 11 | コ | ||
| 12 | サ | ||
| 13 | シ | ||
| 14 | ス | ||
| 15 | セ | ||
| 16 | ソ | ||
| 17 | タ | ||

Identified targeted genes in 4, c, h, イ, カ, セ
Miniprep
| Sample | |
|---|---|
| Host | |
| 4 | |
| C | |
| H | |
| イ | |
| カ | |
| セ | |
Restriction Digest
| Sample | Enzyme | Buffer |
|---|---|---|
| Host | ||
| 4 | Spe1, Xho1 | Buffer H |
| C | Spe1, Xho1 | Buffer H |
| H | Spe1, Xho1 | Buffer H |
| イ | Spe1, Xho1 | Buffer H |
| カ | Spe1, Xho1 | Buffer H |
AGE
| Lane | Sample | Primer | Enzyme | ||
|---|---|---|---|---|---|
| Marker | Host | Insert | |||
| 1 | 1kb Ladder | ||||
| 2 | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | |||
| 3 | 4 | Spe1, Xho1 | |||
| 4 | 4 | Spe1, Xho1 | |||
| 5 | 4 | N.D | |||
| 6 | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | |||
| 7 | c | Spe1, Xho1 | |||
| 8 | c | Spe1, Xho1 | |||
| 9 | c | N.D | |||
| 10 | h | Spe1, Xho1 | |||
| 11 | h | Spe1, Xho1 | |||
| 12 | h | N.D | |||

Transformation (electroportation)
| Host | Sample | |||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| BY4247 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | |
| BY4247 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | |
June 30, 2014
SOGA
Inoculation
Divide each of 2 plates into 8 sections and inoculate following samples on them.
| Host | Sample | Section | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| BY4247 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | A~H |
| BY4247 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | 一~ハ |
July
July 1, 2014
KOBAYASHI
Protein Extraction (Saccharomyces cerevisiae)
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| BY4247 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R |
| BY4247 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R |
July 2, 2014
ONISHI
Miniprep
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| BY4247 | p427-TEF | ||
| BY4247 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R |
| BY4247 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R |
Restriction Digest
| Sample | Enzyme | Buffer | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| p427-TEF | CuMTSE1 | EcoR1 | Buffer H | |
| p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | EcoR1 | Buffer H |
| p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | EcoR1 | Buffer H |
| p427-TEF | CuMTSE1 | Sac1, Kpn1 | Buffer L | |
| p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Sac1, Kpn1 | Buffer L |
| p427-TEF | CuMTSE1 | pGEX6P-2F2,pGEX6P-2R | Sac1, Kpn1 | Buffer L |
AGE
| Lane | Sample | Enzyme | |||
|---|---|---|---|---|---|
| Marker | Vector | Insert | Primer | ||
| 1 | p427-TEF | N.D | |||
| 2 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | N.D | |
| 3 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | N.D | |
| 4 | 1kb Ladder | ||||
| 5 | p427-TEF | EcoR1 | |||
| 6 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | EcoR1 | |
| 7 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | EcoR1 | |
| 8 | 1kb Ladder | ||||
| 9 | p427-TEF | Kpn1 | |||
| 10 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Kpn1 | |
| 11 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Kpn1 | |
| 12 | 1kb Ladder | ||||
| 13 | p427-TEF | Sac1 | |||
| 14 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Sac1 | |
| 15 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Sac1 | |

Restriction Digest
| Sample | Enzyme | Buffer | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| p427-TEF | CuMTSE1 | EcoR1 | Buffer H | |
| p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | EcoR1 | Buffer H |
| p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | EcoR1 | Buffer H |
| p427-TEF | CuMTSE1 | Sac1 | Buffer L | |
| p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Sac1 | Buffer L |
| p427-TEF | CuMTSE1 | pGEX6P-2F2,pGEX6P-2R | Sac1 | Buffer L |
| p427-TEF | CuMTSE1 | Kpn1 | Buffer L | |
| p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Kpn1 | Buffer L |
| p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Kpn1 | Buffer L |
| p427-TEF | CuMTSE1 | Spe1, Xho1 | Buffer H | |
| p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Buffer H |
| p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Spe1, Xho1 | Buffer H |
AGE
| Lane | Sample | Enzyme | |||
|---|---|---|---|---|---|
| Marker | Vector | Insert | Primer | ||
| 1 | 1kb Ladder | ||||
| 2 | p427-TEF | CuMTSE1 | EcoR1 | ||
| 3 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | EcoR1 | |
| 4 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | EcoR1 | |
| 5 | p427-TEF | CuMTSE1 | Kpn1 | ||
| 6 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Kpn1 | |
| 7 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Kpn1 | |
| 8 | p427-TEF | CuMTSE1 | Sac1 | ||
| 9 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Sac1 | |
| 10 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Sac1 | |
| 11 | p427-TEF | CuMTSE1 | Spe1, Xho1 | ||
| 12 | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | |
| 13 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R | Spe1, Xho1 | |
July 3, 2014
OKAMOTO
PCR
| Sample | DNA Polymerase | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| pGEX6P-2 | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTS3 | pGEX6P-2F2, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTS62 | pGEX6P-2F2, pGEX6P-2R | KOD-FX-NEO |
Protein Extraction (Saccharomyces cerevisiae)
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| BY4247 | p427-TEF | ||
| A | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R |
| 六 | p427-TEF | CuMTSE1 | pGEX6P-2F2, pGEX6P-2R |
July 4, 2014
SHIMADA
AGE
| Lane | Sample/Marker | |||
|---|---|---|---|---|
| Marker | Vector | Insert | Primer | |
| 1 | 1kb Ladder | |||
| 2 | pGEX6P-2 | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | |
| 3 | pGEX6P-2 | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | |
| 4 | pGEX6P-2 | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | |
| 5 | pGEX6P-2 | CuMTS3 | pGEX6P-2F2, pGEX6P-2R | |
| 6 | pGEX6P-2 | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | |
| 7 | pGEX6P-2 | CuMTS62 | pGEX6P-2F2, pGEX6P-2R | |

PCR
| Sample | DNA Polymerase | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| pGEX6P-2 | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTS3 | pGEX6P-2F2, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | KOD-FX-NEO |
| pGEX6P-2 | CuMTS62 | pGEX6P-2F2, pGEX6P-2R | KOD-FX-NEO |
AGE
| Lane | Sample | |||
|---|---|---|---|---|
| Marker | Vector | Insert | Primer | |
| 1 | 1kb Ladder | |||
| 2 | pGEX6P-2 | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | |
| 3 | pGEX6P-2 | 1kb Ladder+K83:N91 | pGEX6P-2F2, pGEX6P-2R | |
| 4 | pGEX6P-2 | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | |
| 5 | pGEX6P-2 | CuMTS3 | pGEX6P-2F2, pGEX6P-2R | |
| 6 | pGEX6P-2 | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | |
| 7 | pGEX6P-2 | CuMTS62 | pGEX6P-2F2, pGEX6P-2R | |

July 5, 2014
KOBAYASHI
SDS-PAGE
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| BL21(DE3)pLysS | pGEX6P-2 | ||
| 4 | pGEX6P-2 | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R |
| C | pGEX6P-2 | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R |
| BY4247 | p427-TEF | ||
| A | p427-TEF | CuMTSE1 | pGEX6P-2F1, pGEX6P-2R |
| 六 | p427-TEF | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R |
July 6, 2014
SOGA
DNA Refinement1
| Sample | |
|---|---|
| DNA | Primer |
| CuMTSE2 | pGEX6P-2F1, pGEX6P-2R |
| CuMTSE2 | pGEX6P-2F2, pGEX6P-2R |
| CuMTS3 | pGEX6P-2F1, pGEX6P-2R |
| CuMTS62 | pGEX6P-2F1, pGEX6P-2R |
Restriction Digest
| Vector | Sample | Enzyme | Buffer | |
|---|---|---|---|---|
| DNA | Primer | |||
| CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer | |
| CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer | |
| CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer | p427-TEF | Spe1, Xho1 | Fast digest buffer |
DNA Refinement2
| Sample | Enzyme | |
|---|---|---|
| DNA | Primer | |
| CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
| CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | Spe1, Xho1 |
Ligation
| Vector | Sample | Enzyme | |
|---|---|---|---|
| DNA | Primer | ||
| p427-TEF | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
| p427-TEF | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | Spe1, Xho1 |
DNA ligase: DNA Ligation Kit ‹Mighty Mix› (TAKARA BIO INC.)
Transformation (E.coli)
| Host | Sample | Enzyme | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| DH5α | p427-TEF | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
| DH5α | p427-TEF | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | Spe1, Xho1 |
| DH5α | p427-TEF | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
| DH5α | p427-TEF | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
July 7, 2014
KOBAYASHI
Colony Isolation
Isolate each colony from the plates which were inoculated on July 6.
| Host | Sample | Section | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| DH5α | p427-TEF | CuMTSE2 | pGEX6P-2F1, pGEX6P-2R | 1~16 |
| DH5α | p427-TEF | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | ア~エ |
| DH5α | p427-TEF | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | a~p |
Transformation (E.coli)
| Host | Sample | Enzyme | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| DH5α | p427-TEF | CuMTSE2 | pGEX6P-2F2, pGEX6P-2R | Spe1, Xho1 |
July 8, 2014
OKAMOTO
Colony Sweep
| Lane | Sample | |
|---|---|---|
| Host | ||
| 1 | DH5α | p427-TEF |
| 2 | a | |
| 3 | b | |
| 4 | c | |
| 5 | d | |
| 6 | e | |
| 7 | f | |
| 8 | g | |
| 9 | h | |
| 10 | i | |
| 11 | j | |
| 12 | k | |
| 13 | l | |
| 14 | m | |
| 15 | n | |
| 16 | o | |
| 17 | p | |

Colony Sweep
| Lane | Sample | |
|---|---|---|
| Host | ||
| 1 | DH5α | p427-TEF |
| 2 | ア | |
| 3 | イ | |
| 4 | ウ | |
| 5 | エ | |

Colony Sweep
| Lane | Sample | |
|---|---|---|
| Host | ||
| 1 | DH5α | p427-TEF |
| 2 | 1 | |
| 3 | 2 | |
| 4 | 3 | |
| 5 | 4 | |
| 6 | 5 | |
| 7 | 6 | |
| 8 | 7 | |
| 9 | 8 | |
| 10 | 9 | |
| 11 | 10 | |
| 12 | 11 | |
| 13 | 12 | |
| 14 | 13 | |
| 15 | 14 | |
| 16 | 15 | |
| 17 | 16 | |

July 9, 2014
OKAMOTO
Main Culture
| Sample |
|---|
| Host |
| d |
| h |
| l |
| ア |
| イ |
| エ |
Mini Prep
| Sample |
|---|
| Host |
| D |
| H |
| L |
| ア |
| イ |
| エ |
Restriction Digest
| Vector | Sample | Enzyme | Buffer | |
|---|---|---|---|---|
| Insert | Primer | |||
| p427-TEF(d) | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer |
| p427-TEF(h) | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer |
| p427-TEF(l) | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer |
| p427-TEF(ア) | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer |
| p427-TEF(イ) | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer |
| p427-TEF(エ) | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 | Fast digest buffer |
AGE
| Lane | Sample | Enzyme | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| 1 | p427-TEF(ア) | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | N.D |
| 2 | p427-TEF(ア) | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
| 3 | p427-TEF(イ) | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | N.D |
| 4 | p427-TEF(イ) | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
| 5 | p427-TEF(エ) | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | N.D |
| 6 | p427-TEF(エ) | CuMTS3 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
| 7 | p427-TEF(d) | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | N.D |
| 8 | p427-TEF(d) | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
| 9 | p427-TEF(h) | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | N.D |
| 10 | p427-TEF(h) | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |
| 11 | p427-TEF(l) | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | N.D |
| 12 | p427-TEF(l) | CuMTS62 | pGEX6P-2F1, pGEX6P-2R | Spe1, Xho1 |

August
August 19, 2014
Conducted mutagenesis to remove an illegal restriction site (EcoR1)
InversePCR
| Sample | |||
|---|---|---|---|
| Vector | Insert | Primer | DNA polymer-ase |
| pGEX6P-2 | CuMTS3 | Ter-pinene3-mutagenesis-FP, Terpinene3-mutagenesis-RP | KOD-Plus |
August 20, 2014
Restarted mutagenesis from protocol No.3 to obtain plasmid (muta-CuMTS3 in pGEX-6P-2)
Transformation
| Sample | |||
|---|---|---|---|
| Host | Vector | Insert | Primer |
| DH5α | pGEX6P-2 | Mu-ta-CuMTS3 | Terpinene3-mutagenesis-FP and |
| Terpinene3-mutagenesis-RP | |||
August 21, 2014
Colony Isolation
Isolated each colony from two plates which were inoculated overnight
| Sample | ||||
|---|---|---|---|---|
| Host | Vector | Insert | Primer | Section |
| DH5α | pSB1C3 | Mu-ta-CuMTS3 | Terpinene3-mutagenesis-FP, Terpinene3-mutagenesis-RP |
あ-み |
August 22, 2014
Restriction Digest
| Sample | ||||
|---|---|---|---|---|
| Host | Vector | Insert | Enzyme | Buffer |
| DH5α あ | pGEX6P-2 | Mu-ta-CuMTS3 | Sal1 | Fast Di-gest buffer |
| DH5α か | pGEX6P-2 | Mu-ta-CuMTS3 | Sal1 | Fast Di-gest buffer |
| DH5α さ | pGEX6P-2 | Mu-ta-CuMTS3 | Sal1 | Fast Di-gest buffer |
| DH5α た | pGEX6P-2 | Mu-ta-CuMTS3 | Sal1 | Fast Di-gest buffer |
AGE
| Sample | |||||
|---|---|---|---|---|---|
| Lane | Marker | Host | Vector | Insert | Enzyme |
| 1 | 1kb Ladder | ||||
| 2 | DH5α あ | pGEX6P-2 | Muta-CuMTS3 | Sal1 | |
| 3 | DH5α か | pGEX6P-2 | Muta-CuMTS3 | Sal1 | |
| 2 | DH5α さ | pGEX6P-2 | Muta-CuMTS3 | Sal1 | |
| 2 | DH5α た | pGEX6P-2 | Muta-CuMTS3 | Sal1 | |
Restriction Digest
| Sample | ||||
|---|---|---|---|---|
| Host | Vector | Insert | Enzyme | Buffer |
| DH5α か | pGEX6P-2 | CuMTS3 | EcoR1 | Fast Di-gest buffer |
| DH5α か | pGEX6P-2 | Mu-ta-CuMTS3 | EcoR1 | Fast Di-gest buffer |
AGE
| Sample | Sample | ||||
|---|---|---|---|---|---|
| Marker | Host | Vector | Insert | Enzyme | |
| 1 | 1kb Ladder | ||||
| 2 | DH5α か | pGEX6P-2 | CuMTS3 | EcoR1 | |
| 3 | DH5α か | pGEX6P-2 | Mu-ta-CuMTS3 | EcoR1 | |
PCR
| Sample | |||
|---|---|---|---|
| Vector | Insert | Primer | DNA polymerase |
| か pGEX6P-2 | mu-ta-CuMTS3 | terpinene3-prefix and ter-pinene3-suffix | KOD-FX-Neo-Plus |
AGE
| Sample | |||||
|---|---|---|---|---|---|
| Lane | Marker | Vector | Insert | Primer | DNA polymerase |
| 1 | 1kb Ladder | ||||
| 2 | か pGEX6P-2 | mu-ta-CuMTS3 | terpinene3-prefix and ter-pinene3-suffix | KOD-FX-Neo-Plus | |
August 30, 2014
PCR| Sample | |||
|---|---|---|---|
| Vector | Insert | Primer | DNA polymerase |
| か pGEX6P-2 | mu-ta-CuMTS3 | terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1 | Taq |
AGE
| Sample | ||||
|---|---|---|---|---|
| Lane | Marker | Vector | Insert | Primer |
| 1 | 1kb Ladder | |||
| 2 | か pGEX6P-2 | mu-ta-CuMTS3 | terpinene3-prefix-Xba1 and ter-pinene3-suffix-Spe1 | |
Made total amunt of 295 μL of PCR samples.
September
September 18, 2014
Miniprep| Sample | ||
|---|---|---|
| Host | Vector | Insert |
| DH5α | pSB1C3 | BBa_J04450 |
| DH5α | pSB1C3 | BBa_J04450 |
| DH5α | pSB1C3 | BBa_J04450 |
Restriction Digest
| Sample | |||
|---|---|---|---|
| Vector | Insert | Enzyme | Buffer |
| pSB1C3 | BBa_J04450 | EcoR1, Pst1 | Buffer H |
| pSB1C3 | BBa_J04450 | Xba1, Spe1 | Fast Digest buffer |
| pSB1C3 | BBa_J04450 | ||
AGE
| Sample | |||||
|---|---|---|---|---|---|
| Lane | Marker | Host | Vector | Insert | Enzyme |
| 1 | 1kb Ladder | /td> | |||
| 2 | DH5α | pSB1C3 | BBa_J04450 | EcoR1, Pst1/td> | |
| 3 | DH5α | pSB1C3 | BBa_J04450 | Xba1, Spe1 | |
| 2 | DH5α | pSB1C3 | BBa_J04450 | /td> | |
PCR
| Sample | |||
|---|---|---|---|
| Vector | Insert | Primer | DNA polymerase |
| か pGEX6P-2 | mu-ta-CuMTS3 | terpinene3-prefix-Xba1 and ter-pinene3-suffix-Spe1 | Taq |
terpinene3-prefix-Xba1 and ter-pinene3-suffix-Spe1
| Sample | |
|---|---|
| Insert | Primer |
| mu-ta-CuMTS3 | terpinene3-prefix-Xba1 and ter-pinene3-suffix-Spe1 |
Restriction Digest
| Sample | ||||
|---|---|---|---|---|
| Vector | Insert | Primer | Enzyme | Buffer |
| pSB1C3 | Xba1, Spe1 | Fast Digest | ||
| かpGEX6P-2 | muta-CuMTS3 | terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1 | Xba1, Spe1 | Fast Digest |
DNA Refinement 2
| Sample | |
|---|---|
| Insert | Primer |
| muta-CuMTS3 | terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1 |
Ligation
| Sample | |||
|---|---|---|---|
| Vector | DNA | Primer | Enzyme |
| pSB1C3 | muta-CuMTS3 | terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1 | Spe1, Xba1 |
DNA ligase: DNA Ligation Kit ‹Mighty Mix› (TAKARA BIO INC.)
Transformation
| Sample | |||
|---|---|---|---|
| Host | Vector | Insert | Primer |
| DH5α | pSB1C3 | muta-CuMTS3 | terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1 |
September 19, 2014
Colony IsolationIsolate each colony from the plates which were inoculated overnight.
| Sample | ||||
|---|---|---|---|---|
| Host | Vector | Insert | Primer | section |
| DH5α | pSB1C3 | mu-ta-CuMTS3 | terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1 | A-P |
| DH5α | pSB1C3 | mu-ta-CuMTS3 | terpinene3-prefix-Xba1 and terpinene3-suffix-Spe1 | ア-タ |
Colony Sweep
| Lane | Sample | ||
|---|---|---|---|
| Host | Vector | Insert | |
| 1 | DH5α | pSB1C3 | Muta-CuMTS3 |
| 2 | ア | ||
| 3 | イ | ||
| 4 | ウ | ||
| 5 | エ | ||
| 6 | オ | ||
| 7 | カ | ||
| 8 | キ | ||
| 9 | ク | ||
| 10 | ケ | ||
| 11 | コ | ||
| 12 | サ | ||
| 13 | シ | ||
| 14 | ス | ||
| 15 | セ | ||
| 16 | ソ | ||
| 17 | タ | ||
Targeted colonies were identified in セand ソ.
Colony Sweep
| Lane | Sample | ||
|---|---|---|---|
| Host | Vector | Insert | |
| 1 | DH5α | pSB1C3 | Muta-CuMTS3 |
| 2 | A | ||
| 3 | B | ||
| 4 | C | ||
| 5 | D | ||
| 6 | E | ||
| 7 | F | ||
| 8 | G | ||
| 9 | H | ||
| 10 | I | ||
| 11 | J | ||
| 12 | K | ||
| 13 | L | ||
| 14 | M | ||
| 15 | N | ||
| 16 | O | ||
| 17 | P | ||
Targeted colonies were identified in colony N and F.
Selected セ, ソ, N, F
Main Culture
| Sample | |||
|---|---|---|---|
| Host | Vector | Insert | Primer |
| DH5α(N) | pSB1C3 | mu-ta-CuMTS3 | ---------- |
| DH5α(F) | pSB1C3 | mu-ta-CuMTS3 | ---------- |
| DH5α(セ) | pSB1C3 | mu-ta-CuMTS3 | ---------- |
| DH5α(ソ) | pSB1C3 | mu-ta-CuMTS3 | ---------- |
Cultivated these samples in 3mL of LB medium with ChlPhe at 30°C overnight.
September 20, 2014
Miniprep| Sample | ||
|---|---|---|
| Host | Vector | Primer |
| DH5α(N) | pSB1C3 | Mu-ta-CuMTS3 |
| DH5α(F) | pSB1C3 | Mu-ta-CuMTS3 |
| DH5α(セ) | pSB1C3 | Mu-ta-CuMTS3 |
| DH5α(ソ) | pSB1C3 | Mu-ta-CuMTS3 |
Enzyme: E/P and Xba1
Restriction Digest
| Sample | Enzyme | Buffer | |||
|---|---|---|---|---|---|
| Marker | Host | Vector | Insert | ||
| pSB1C3 | |||||
| DH5α (セ) | pSB1C3 | Muta-CuMTS3 | |||
| DH5α (ソ) | pSB1C3 | Muta-CuMTS3 | |||
| DH5α (F) | pSB1C3 | Muta-CuMTS3 | |||
| DH5α (N) | pSB1C3 | Muta-CuMTS3 | |||
| DH5α (セ) | pSB1C3 | EcoR1, Pst1 | Buffer H | ||
| DH5α (ソ) | pSB1C3 | Muta-CuMTS3 | EcoR1, Pst1 | Buffer H | |
| DH5α (F) | pSB1C3 | Muta-CuMTS3 | EcoR1, Pst1 | Buffer H | |
| DH5α (N) | pSB1C3 | Muta-CuMTS3 | EcoR1, Pst1 | Buffer H | |
| pSB1C3 | Muta-CuMTS3 | EcoR1, Pst1 | Buffer H | ||
| pSB1C3 | Xba1 | FD buffer | |||
| DH5α (セ) | pSB1C3 | Muta-CuMTS3 | Xba1 | FD buffer | |
| DH5α (ソ) | pSB1C3 | Muta-CuMTS3 | Xba1 | FD buffer | |
| DH5α (F) | pSB1C3 | Muta-CuMTS3 | Xba1 | FD buffer | |
| DH5α (N) | pSB1C3 | Muta-CuMTS3 | Xba1 | FD buffer | |
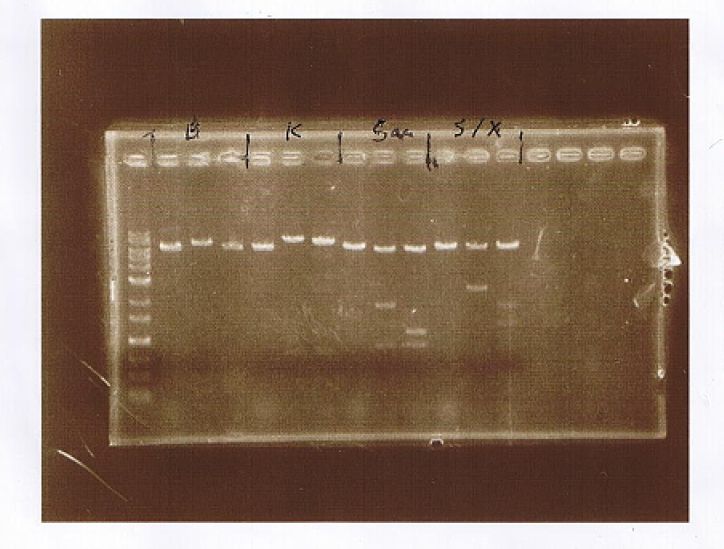
AGE
| Lane | Sample | Enzyme | |||
|---|---|---|---|---|---|
| Marker | Host | Vector | Insert | ||
| 1 | pSB1C3 | ||||
| 2 | DH5α (セ) | pSB1C3 | Muta-CuMTS3 | ||
| 3 | DH5α (ソ) | pSB1C3 | Muta-CuMTS3 | ||
| 4 | DH5α (F) | pSB1C3 | Muta-CuMTS3 | ||
| 5 | DH5α (N) | pSB1C3 | Muta-CuMTS3 | ||
| 6 | 1kb Ladder | ||||
| 7 | DH5α (セ) | pSB1C3 | Muta-CuMTS3 | EcoR1, Pst1 | |
| 8 | DH5α (ソ) | pSB1C3 | Muta-CuMTS3 | EcoR1, Pst1 | |
| 9 | DH5α (F) | pSB1C3 | Muta-CuMTS3 | EcoR1, Pst1 | |
| 10 | DH5α (N) | pSB1C3 | Muta-CuMTS3 | EcoR1, Pst1 | |
| 11 | pSB1C3 | Muta-CuMTS3 | EcoR1, Pst1 | ||
| 12 | 1kb Ladder | ||||
| 13 | pSB1C3 | Xba1 | |||
| 14 | DH5α (セ) | pSB1C3 | Muta-CuMTS3 | Xba1 | |
| 15 | DH5α (ソ) | pSB1C3 | Muta-CuMTS3 | Xba1 | |
| 16 | DH5α (F) | pSB1C3 | Muta-CuMTS3 | Xba1 | |
| 17 | DH5α (N) | pSB1C3 | Muta-CuMTS3 | Xba1 |
Sample F was selected as a submission plasmid.
Main Culture
| Sample | |||
|---|---|---|---|
| Host | Vector | Insert | Primer |
| DH5α(F) | pSB1C3 | mu-ta-CuMTS3 | ---------- |
| DH5α(F) | pSB1C3 | mu-ta-CuMTS3 | ---------- |
| DH5α(F) | pSB1C3 | mu-ta-CuMTS3 | ---------- |
Cultivated these samples in 3mL of LB medium with ChlPhe at 30°C overnight.
September 21, 2014
Miniprep| Sample | |||
|---|---|---|---|
| Host | Vector | Insert | Primer |
| DH5α(F) | pSB1C3 | Mu-ta-CuMTS3 | |
| DH5α(F) | pSB1C3 | Mu-ta-CuMTS3 | |
| DH5α(F) | pSB1C3 | Mu-ta-CuMTS3 | |
Restriction Digest
| Sample | Enzyme | Buffer | ||||
|---|---|---|---|---|---|---|
| Marker | Host | Vector | Insert | Primer | ||
| 1kb Ladder | ||||||
| pSB1C3 | BBa_J04450 | EcoR1 | Buffer H | |||
| pSB1C3 | BBa_J04450 | Pst1 | Buffer H | |||
| pSB1C3 | BBa_J04450 | EcoR1, Pst1 | Buffer H | |||
| pSB1C3 | BBa_J04450 | Spe1 | FD buffer | |||
| pSB1C3 | BBa_J04450 | Xba1 | FD buffer | |||
| pSB1C3 | BBa_J04450 | Spe1, Xba1 | FD buffer | |||
| pSB1C3 | BBa_J04450 | |||||
| λDNA-HindⅢ | ||||||
| F | pSB1C3 | Mu-ta-CuMTS3 | EcoR1 | Buffer H | ||
| F | pSB1C3 | Mu-ta-CuMTS3 | Pst1 | Buffer H | ||
| F | pSB1C3 | Mu-ta-CuMTS3 | EcoR1, Pst1 | Buffer H | ||
| F | pSB1C3 | Mu-ta-CuMTS3 | Spe1 | FD buffer | ||
| F | pSB1C3 | Mu-ta-CuMTS3 | Xba1 | FD buffer | ||
| F | pSB1C3 | Mu-ta-CuMTS3 | Spe1, Xba1 | FD buffer | ||
| F | pSB1C3 | Mu-ta-CuMTS3 | ||||
AGE
| Lane | Sample | Enzyme | |||
|---|---|---|---|---|---|
| Marker | Host | Vector | Insert | ||
| 1 | pSB1C3 | ||||
| 2 | DH5α (セ) | pSB1C3 | Muta-CuMTS3 | ||
| 3 | DH5α (ソ) | pSB1C3 | Muta-CuMTS3 | ||
| 4 | DH5α (F) | pSB1C3 | Muta-CuMTS3 | ||
| 5 | DH5α (N) | pSB1C3 | Muta-CuMTS3 | ||
| 6 | 1kb Ladder | ||||
| 7 | DH5α (セ) | pSB1C3 | Muta-CuMTS3 | EcoR1, Pst1 | |
| 8 | DH5α (ソ) | pSB1C3 | Muta-CuMTS3 | EcoR1, Pst1 | |
| 9 | DH5α (F) | pSB1C3 | Muta-CuMTS3 | EcoR1, Pst1 | |
| 10 | DH5α (N) | pSB1C3 | Muta-CuMTS3 | EcoR1, Pst1 | |
| 11 | pSB1C3 | Muta-CuMTS3 | EcoR1, Pst1 | ||
| 12 | 1kb Ladder | ||||
| 13 | pSB1C3 | Xba1 | |||
| 14 | DH5α (セ) | pSB1C3 | Muta-CuMTS3 | Xba1 | |
| 15 | DH5α (ソ) | pSB1C3 | Muta-CuMTS3 | Xba1 | |
| 16 | DH5α (F) | pSB1C3 | Muta-CuMTS3 | Xba1 | |
| 17 | DH5α (N) | pSB1C3 | Muta-CuMTS3 | Xba1 |
Sample F was selected as a submission plasmid.
Main Culture
| Sample | |||
|---|---|---|---|
| Host | Vector | Insert | Primer |
| DH5α(F) | pSB1C3 | mu-ta-CuMTS3 | ---------- |
| DH5α(F) | pSB1C3 | mu-ta-CuMTS3 | ---------- |
| DH5α(F) | pSB1C3 | mu-ta-CuMTS3 | ---------- |
Cultivated these samples in 3mL of LB medium with ChlPhe at 30°C overnight.
a
a
a
a
a
a
a
a
a
a
a
a
a
a
a
October
October 1, 2014
Protein Extraction (S.cerevisiae)| Sample | |||
|---|---|---|---|
| Host | Vector | Insert | Primer |
| BY4247 | p427-TEF | ||
| BY4247 | p427-TEF | CuMTSE2 | P-F1, P-R/td> |
| BY4247 | p427-TEF | CuMTS3 | P-F2, P-R |
| BY4247 | p427-TEF | CuMTS62 | P-F2, P-R |
SDS-PAGE
| Lane | Sample | |||
|---|---|---|---|---|
| Host | Vector | Insert | Primer | |
| 1 | BY4247 | p427-TEF | ||
| 2 | BY4247 | p427-TEF | CuMTSE2 | P-F1, P-R |
| 3 | BY4247 | p427-TEF | CuMTS3 | P-F2, P-R |
| 4 | BY4247 | p427-TEF | CuMTS62 | P-F2, P-R |
October 2, 2014
Preculture| Sample | |||
|---|---|---|---|
| Host | Vector | Insert | Primer |
| BY4247 | p427-TEF | ||
| BY4247 | p427-TEF | CuMTS3 | P-F2, P-R |
Western blot
| Lane | Sample | |||
|---|---|---|---|---|
| Host | Vector | Insert | Primer | |
| 1 | BY4247 | p427-TEF | ||
| 2 | BY4247 | p427-TEF | CuMTSE2 | P-F1, P-R |
| 3 | BY4247 | p427-TEF | CuMTS3 | P-F2, P-R |
| 4 | BY4247 | p427-TEF | CuMTS62 | P-F2, P-R |
October 3, 2014
Main cultureCultivated following samples in Big Scale with G418 at 28°C overnight in a shaken culture.
| Sample | |||
|---|---|---|---|
| Host | Vector | Insert | Primer |
| BY4247 | p427-TEF | ||
| BY4247 | p427-TEF | CuMTS3 | P-F2, P-R |
Restriction Digest
| Sample | Enzyme | Buffer | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| pSB1C3 | EcoR1, Spe1 | Buffer H | ||
| pSB1C3 | Xba1, Pst1 | Buffer H | ||
| Linearized pSB1A3 | EcoR1, Pst1 | Buffer H | ||
AGE
| Lane | Marker | Sample | Enzyme | ||
|---|---|---|---|---|---|
| Vector | Insert | Primer | |||
| 1 | 1kb Ladder | ||||
| 2 | pSB1C3 | EcoR1, Spe1 | |||
| 3 | pSB1C3 | Xba1, Pst1 | |||
| 4 | Linearized pSB1A3 | EcoR1, Pst1 | |||
DNA Refinement 1
| Sample | Enzyme | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| pSB1C3 | EcoR1, Spe1 | ||
| pSB1C3 | Xba1, Pst1 | ||
| pSB1C3 | Xba1, Pst1 | ||
| Linearized pSB1A3 | EcoR1, Pst1, Dpn1 | ||
Ligation
| Vector | Sample | Enzyme | |
|---|---|---|---|
| Insert1 | Insert2 | ||
| pSB1A3 | EcoR1, Xba1, Spe1, Pst1 | ||
| pSB1A3 | EcoR1, Xba1, Spe1, Pst1 | ||
DNA ligase: DNA Ligation Kit ‹Mighty Mix› (TAKARA BIO INC.)
Transformation
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert1 | Insert2 | |
| DH5α | pSB1A3 | ||
| DH5α | pSB1A3 | ||
October 4, 2014
Colony IsolationIsolate each colony from the plates which were inoculated overnight.
| Host | Sample | Section | ||
|---|---|---|---|---|
| Vector | Insert | Primer | ||
| DH5α | pSB1A3 | 1-16 | ||
| DH5α | pSB1A3 | a-p | ||
Monoterpene Extraction
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| BY4247 | p427-TEF | ||
| BY4247 | p427-TEF | CuMTS3 | P-F2, P-R |
Samples were stocked in -4℃.
Preculture
| Host | Sample | ||
|---|---|---|---|
| Vector | Insert | Primer | |
| BY4247 | p427-TEF | CuMTSE2 | P-F2, P-R |
| BY4247 | p427-TEF | CuMTS62 | P-F2, P-R |
Cultivated samples in 3mL of SD medium with G418 at 30°C overnight.
October 5, 2014
Miniprep| Host | Sample | |
|---|---|---|
| Vector | Insert | |
| DH5α | pSB1A3 | |
| DH5α | pSB1A3 | |
Restriction Digest and AGE
| Host | Sample | Enzyme | |
|---|---|---|---|
| Vector | Insert | ||
| DH5α | pSB1C3 | EcoR1, Pst1 | |
| DH5α | pSB1C3 | EcoR1, Pst1 | |
| pSB1C3 | EcoR1, Pst1 | ||
a
a
a
a
a
a
a
a
a
a
a
a
a
a
a
a
a
a
a
a
a
a
a
a
a
 "
"
