Team:Evry/Interlab Study
From 2014.igem.org
| (14 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
{{:Team:Evry/Template:Menu}} | {{:Team:Evry/Template:Menu}} | ||
{{:Team:Evry/Template:InterlabstudyTop}} | {{:Team:Evry/Template:InterlabstudyTop}} | ||
| + | |||
<html> | <html> | ||
| - | < | + | <div id="tab-circle"class="row about-details"> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <div class="col-md-4 col-sm-4 col-xs-6"> | ||
| + | <div class="animated hiding" data-animation="fadeInDown" data-delay="0"> | ||
| + | <FONT color="white"> <h5>Chassis</h5></font> | ||
| + | |||
| + | <a href="#chassis" role="tab" data-toggle="tab"><img class="img-circle" src="https://static.igem.org/mediawiki/2014/7/7e/Chassis_EVRY.png" height="128" width="128" alt="Conference" border="30px" style="box-shadow: 0 0 8px rgba(0, 0, 0, .8); "/></a> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="col-md-4 col-sm-4 col-xs-6"> | ||
| + | <div class="animated hiding" data-animation="fadeInDown" data-delay="500"> | ||
| + | <font colors ="white"><h5>Transposons</h5></font> | ||
| + | <a href="#transposons" role="tab" data-toggle="tab"><img class="img-circle" src="https://static.igem.org/mediawiki/2014/b/bf/Transpo3.png" height="128" width="128" alt="Workshops" style="box-shadow: 0 0 8px rgba(0, 0, 0, .8); "></a> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="clearfix visible-xs"></div> | ||
| + | <div class="col-md-4 col-sm-4 col-xs-6"> | ||
| + | <div class="animated hiding" data-animation="fadeInDown" data-delay="1000"> | ||
| + | <FONT color="white"><h5>Target</h5></font> | ||
| + | <a href="#target" role="tab" data-toggle="tab"><img class="img-circle" src="https://static.igem.org/mediawiki/2014/f/f1/Image1.jpg" height="128" width="128" alt="Hackathons" style="box-shadow: 0 0 8px rgba(0, 0, 0, .8)"></a> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| - | + | </section> | |
| - | {{:Team:Evry/Interlab_Study/ | + | <div class="tab-content"> |
| - | {{:Team:Evry/Interlab_Study/ | + | <div class="tab-pane active" id="chassis"></html>{{:Team:Evry/Interlab_Study/Results}}<html></div> |
| - | {{:Team:Evry/Interlab_Study/ | + | <div class="tab-pane" id="transposons"></html>{{:Team:Evry/Interlab_Study/Used_Devices}}<html></div> |
| - | + | <div class="tab-pane" id="target"></html>{{:Team:Evry/Interlab_Study/Notebook}}<html></div> | |
| - | + | </div> | |
| - | + | <script> | |
| - | + | $('#tab-circle a').click(function (e) { | |
| - | + | e.preventDefault() | |
| - | + | $(this).tab('show') | |
| - | + | }) | |
| - | + | </script> | |
| - | + | <!--##########################--> | |
| - | + | <section id="statistic" class="parallax" style="background-image: url(https://static.igem.org/mediawiki/2014/4/45/Under-water-wallpaper-hd-104.jpg);"> | |
| + | <div class="overlay"></div> | ||
| + | </html> | ||
{{:Team:Evry/Template:HomeFooter}} | {{:Team:Evry/Template:HomeFooter}} | ||
Latest revision as of 21:12, 17 October 2014
Interlab study
Interlab study - Aim & Results
There were two sub-parts in this study:
- The mandatory one: obtain fluorescence data for three specific genetic devices expressing GFP and compare them.
- The Extra Credit assignments: we choose to study the entire Anderson library of constitutive prokaryotic promoters (J23100 to J23119).
This library corresponds to 19 constitutive promoters containing some nucleotide mutations on the -35 and the -10, as shown Used Biobricks and plasmids . The idea is to compare the expression strength of promoters according to mutations, from the maximum amount of fluorescence data in order to have significant results.
Required Devices
 |
 |
Theoritically constructions I20260 and J3101 were similar, constructions and alignments in sillico matched perfectly. The difference between these constructions lies on the assembly way. However we obtained, as some other teams (University of Oxford, Paris-Bettencourt and ITESM-CEM), significant GFP expression difference. There is probably a Biobrick standard construction problem, which relates to the traited question on our Phylosophy subsection.
Entire Anderson library of constitutive promoters (J23100-J23119)
 |
 |
Unfortunatly, we did not collect satisfying data for the 19 constructions,although assays were repeated several times. The data profile was repeatable under people, apparatus and strain. Strain carrying promotorless vector seemed to had higher fluorescence intensity than strain containing constructions from the Anderson library, that explains negative curve on Corrected GFP fluorescence intensity according to OD 600 nm graph. However exploitable data were collected for 5 constructions (100, 104, 105, 107 and 118) for which the relationship between corrected GFP fluorescence and OD 600 nm was linear that corresponded to the expected profile for a constitutive promoter (See graph Corrected GFP fluorescence intensity according to OD 600 nm). Corrected GFP fluorescence intensity at OD 600 nm = 0.45 of these 5 constructions were presented on bar chart above and compared with registry data on table below.

Possible approach to overcome data acquisition problems:
- Using M9 medium instead of LB medium to minimize the basal medium fluorescence
- Using another E. coli strain that has a smaller natural fluorescence
- Changing of dilution and gain to find the optimal one
- Choosing a TECAN with a lower detection limits to collect data for weak promoters
Interlab study - Used devices
Construction way

Constructions were built using classical Biobrick restriction sites.
- Required device 1: originally this device was carried by PSB1K3 and was sud-cloned onto PSB1C3.
- Vector: digestion of PSB1C3 and PSB1K3 carrying I20260 via EcoRi and PstI
- Insert: digestion of PSB1K3 carrying I20260 via EcoRi and PstI
- Ligation with T4 DNA ligase (New England Biolabs)
- Required device 2, 3 and the 19 constructions for the entire Anderson library: promoters were inserted into PSB1C3 carrying E0240. Process is described on following figure 1.
- Vector: digestion of PSB1C3 carrying E0240 via EcoRI and XbaI
- Insert: digestion of PSB1C3 carrying K823012* and J61002 carrying J23100 to J23119 via EcoRI and SpeI
- Ligation with T4 DNA ligase (New England Biolabs)
Analysis protocol
Fluorescence data were analyzed via following formula that come from 2010 De Jong et al Experimental and computational validation of models of fluorescent and luminescent reporter genes in bacteria .

Equation B: B(t) is the absorbance of the strain carrying the promoterless vector, Iu(t) the uncorrected fluorescence intensity of DH5 alpha with the functional reporter system and Ib(t) the background fluorescence intensity of LB chloramphenicol medium with the DH5 alpha carrying the promoterless vector.
Used Biobricks and plasmids
Sequencing results
AGTCAGTGAGCGAGGAAGCCTGCATAACGCGAAGTAATCTTTTCGGTTTTAAAGAAAAAGGGCAGGGTGGTGACACCTTGCCCTTTTTTGCCGGACTGCAGCGGCCGCTACTAGTATATAAACGCAGAAAGGCCCACCCGAAGGTGAGCCAGTGTGACTCTAGTAGAGAGCGTTCACCGACAAACAACAGATAAAACGAAAGGCCCAGTCTTTCGACTGAGCCTTTCGTTTTATTTGATGCCTGGCTCTAGTATTATTATTTGTATAGTTCATCCATGCCATGTGTAATCCCAGCAGCTGTTACAAACTCAAGAAGGACCATGTGGTCTCTCTTTTCGTTGGGATCTTTCGAAAGGGCAGATTGTGTGGACAGGTAATGGTTGTCTGGTAAAAGGACAGGGCCATCGCCAATTGGAGTATTTTGTTGATAATGGTCTGCTAGTTGAACGCTTCCATCTTCAATGTTGTGTCTAATTTTGAAGTTAACTTTGATTCCATTCTTTTGTTTGTCTGCCATGATGTATACATTGTGTGAGTTATAGTTGTATTCCAATTTGTGTCCAAGAATGTTTCCATCTTCTTTAAAATCAATACCTTTTAACTCGATTCTATTAACAAGGGTATCACCTTCAAACTTGACTTCAGCACGTGTCTTGTAGTTCCCGTCATCTTTGAAAAATATAGTTCTTTCCTGTACATAACCTTCGGGCATGGCACTCTTGAAAAAGTCATGCTGTTTCATATGATCTGGGTATCTCGCANAGCATTGAACACCATAACCGAAAGTAGTGACAAGTGTTGGCCATGGAACAGGTAGTTTTCCAGTAGTGCAAATAAATTTAAGGGTAAGTTTTCCGTATGTTGCATCACCTTCACCCTCTCCACTGACAGAAAATTTGTGCCCATTAACATCACCATCTAATTCAACAAGAATTGGGACAACTCCAGTGAAAAGTTCTTCTCCTTTACGCATCTAGTACTTTCCTGTGTGACTCTAGTAGCTAGCATAATACCTAGGACTGAGCTNNNTGTAAACTCTNNNANCGGCC
ATAAAANTNGGCGTNTCACGAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTAAGGATGATTTCTGGAATTCGCGGCCGCATCTAGAGTTTACAGCTAGCTCAGTCCTAGGTATTATGCTAGCTTCTAGAGTCACACAGGAAAGTACTAGATGCGTAAAGGAGAAGAACTTTTCACTGGAGTTGTCCCAATTCTTGTTGAATTAGATGGTGATGTTAATGGGCACAAATTTTCTGTCAGTGGAGAGGGTGAAGGTGATGCAACATACGGAAAACTTACCCTTAAATTTATTTGCACTACTGGAAAACTACCTGTTCCATGGCCAACACTTGTCACTACTTTCGGTTATGGTGTTCAATGCTTTGCGAGATACCCAGATCATATGAAACAGCATGACTTTTTCAAGAGTGCCATGCCCGAAGGTTATGTACAGGAAAGAACTATATTTTTCAAAGATGACGGGAACTACAAGACACGTGCTGAAGTCAAGTTTGAAGGTGATACCCTTGTTAATAGAATCGAGTTAAAAGGTATTGATTTTAAAGAAGATGGAAACATTCTTGGACACAAATTGGAATACAACTATAACTCACACAATGTATACATCATGGCAGACAAACAAAAGAATGGAATCAAAGTTAACTTCAAAATTAGACACAACATTGAAGATGGAAGCGTTCAACTAGCAGACCATTATCAACAAAATACTCCAATTGGCGATGGCCCTGTCCTTTTACCAGACAACCATTACCTGTCCACACAATCTGCCCTTTCGAAAGATCCCAACGAANAGAGAGACCACATGGTCCTTCTTGAGTTTGTAACAGCTGCTGGGATTACACATGGCATGGATGAACTATACAAATAATAATACTAGAGCCAGGCATCANATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCGGTGAACGCTCTCTACTAGAGTCACACTGGCTCACCTTCGGGTGGGCCTTTCTGCGTTTATATACTAGTAGCGGCCGCTGCAGTCCGGCAANANAGGGCAAGGTGTCACCACCCTGCCCTTTTTCTTTAAAACCGANAAGATTACTTCGCGTTATGCAGGCTTNCTCGCTCACTGACTCGCTGCGCTCGGTCGTTCGGCTGCGGCGAACGG
TTACTATAAANTAGGCGTATCACGAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTAAGGATGATTTCTGGAATTCGCGGCCGCTTCTAGAGTTTATAGCTAGCTCAGTCCTAGGTACAATGCTAGCTACTAGAGCACACAGGAAAGTACTAGATGCGTAAAGGAGAAGAACTTTTCACTGGAGTTGTCCCAATTCTTGTTGAATTAGATGGTGATGTTAATGGGCACAAATTTTCTGTCAGTGGAGAGGGTGAAGGTGATGCAACATACGGAAAACTTACCCTTAAATTTATTTGCACTACTGGAAAACTACCTGTTCCATGGCCAACACTTGTCACTACTTTCGGTTATGGTGTTCAATGCTTTGCGAGATACCCAGATCATATGAAACAGCATGACTTTTTCAAGAGTGCCATGCCCGAAGGTTATGTACAGGAAAGAACTATATTTTCAAAGATGACGGGAACTACAAGACACGTGCTGAAGTCAAGTTTGAAGNNGATACCCTTGTTAATAGAATCGAGTTAAAAGGTATTGATTTTAAAGAAGATGGAAACATTCTTGGACACANATTGNAATACAACTATAACTCACACAATGTATACATCATGGCAGACAANCNNNAGAATGNAATCAAANTTAACTTCAAAATTANACACAACA
TAGGCGTATNANGAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTAAGGATGATTTCTGGAATTCGCGGCCGCTTCTAGAGTTTACAGCTAGCTCAGTCCTAGGTATTATGCTAGCTACTAGAGTCACACAGGAAAGTACTAGATGCGTAAAGGAGAAGAACTTTTCACTGGAGTTGTCCCAATTCTTGTTGAATTAGATGGTGATGTTAATGGGCACAAATTTTCTGTCAGTGGAGAGGGTGAAGGTGATGCAACATACGGAAAACTTACCCTTAAATTTATTTGCACTACTGGAAAACTACCTGTTCCATGGCCAACACTTGTCACTACTTTCGGTTATGGTGTTCAATGCTTTGCGAGATACCCAGATCATATGAAACAGCATGACTTTTTCAAGAGTGCCATGCCCGAAGGTTATGTACAGGAAAGAACTATATTTTTCAAAGATGACGGGAACTACAAGACACGTGCTGAAGTCAAGTTTGAAGGTGATACCCTTGTTAATAGAATCGAGTTAAAAGGTATTGATTTTAAAGAAGATGGAAACATTCTTGGACACAAATTGGAATACAACTATAACTCACACAATGTATACATCATGGCAGACAAACAAAAGAATGGAATCAAAGTTAACTTCAAAATTAGACACAACATTGAAGATGGAAGCGTTCAACTAGCAGACCATTATCAACAAAATACTCCAATTGGCGATGGCCCTGTCCTTTTACCAGACAACCATTACCTGTCCACACAATCTGCCCTTTCGAAAGATCCCAACGAANAGAGAGACCACATGGTCCTTCTTGAGTTTGTAACAGCTGCTGGGATTACACATGGCATGGATGAACTATACAAATAATAATACTAGAGCCAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCGGTGAACGCTCTCTACTAGAGTCACACTGGCTCACCTTCGGGTGGGCCTTTCTGCGTTTATATACTAGTAGCGGCCGCTGCAGTCCGGCAAAAAAGGGCAAGGTGTCACCACCCTGCCCTTTTTCTTTAAAACCGAAAANATTACTTCNCGTTATGCAGGCTTCCTCGCTCACTGACTCGCTGCGCTCGGTCGTTCGGCTGCGGCGNACGG
Interlab study - Notebook

The three required devices

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23101 - PSB1C3+E0240+J23104
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 80-500nM. (09h00-16h00)

PSB1C3+E0240+J23119:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 80-500nM. (09h00-16h00)

Oct 11

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23110 - PSB1C3+E0240+J23107
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 80-500nM. (09h00-16h00)

PSB1C3+E0240+J23106:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 80-500nM. (09h00-16h00)

PSB1C3+E0240+J23108 - PSB1C3+E0240+J23118 - PSB1C3+E0240+J23111
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 80-500nM. (14h00-21h00)

PSB1C3+E0240+J23102:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 80-500nM. (14h00-21h00)

PSB1C3+E0240+J23101 - PSB1C3+E0240+J23104 - PSB1C3+E0240+J23119:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h at 37°C.
Oct 10

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23109 - PSB1C3+E0240+J23117
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 80-500nM. (11h00-18h00)

PSB1C3+E0240+J23114:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 80-500nM. (11h00-18h00)

PSB1C3+E0240+J23115 - PSB1C3+E0240+J23116
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 80-500nM. (16h00-23h00)

PSB1C3+E0240+J23105:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 80-500nM. (16h00-23h00)

PSB1C3+E0240+J23110 - PSB1C3+E0240+J23107 - PSB1C3+E0240+J23106:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h at 37°C.
PSB1C3+E0240+J23108 - PSB1C3+E0240+J23118 - PSB1C3+E0240+J23111 - PSB1C3+E0240+J23102:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h at 37°C.
Oct 09

Construction n°1: PSB1C3 with I20260
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 80-500nM. (12h00-17h00)

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23100:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 80-500nM. (12h00-17h00)

PSB1C3+E0240+J23112 - PSB1C3+E0240+J23103 - PSB1C3+E0240+J23113:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h at 37°C.
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 80-500nM. (17h00-00h00)

PSB1C3+E0240+J23109 - PSB1C3+E0240+J23117 - PSB1C3+E0240+J23114:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h at 37°C.
PSB1C3+E0240+J23115 - PSB1C3+E0240+J23116 - PSB1C3+E0240+J23105:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h at 37°C.
Oct 08

Construction n°1: PSB1C3 with I20260
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 80-500nM. (12h00-19h00)

Construction n°2: PSB1C3 with J23101-E1010
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 80-500nM. (12h00-19h00)

Construction n°3: PSB1C3 with K823012-E1010
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h at 37°C.
Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23100:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h at 37°C.
Oct 07

Construction n°1: PSB1C3 with I20260
Construction n°2: PSB1C3 with J23101-E1010
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h at 37°C.
Construction n°3: PSB1C3 with K823012-E1010
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h at 37°C.
Other constructions of the Anderson library of constitutive promoters
Oct 06

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23100:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 80-500nM.
OVERFLOW
New pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23107:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 (19h00-02h00).

PSB1C3+E0240+J23109:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP < 80nM.
OVERFLOW
New pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
Oct 04

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23100:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 80-500nM.
OVERFLOW
New pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23107:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 (19h00-02h00).

PSB1C3+E0240+J23109:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP < 80nM.
OVERFLOW
New pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
Oct 04

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23100:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 60-250nM.
OVERFLOW
New pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23105:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 (17h00-00h00).

PSB1C3+E0240+J23107:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23109:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 60-250nM.
OVERFLOW
New pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23117:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 (17h00-00h00).

OVERFLOW
Oct 03

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23100:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 60-100nM.
TECAN FAILED
New pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23105:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23107:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 60-100nM.
OVERFLOW
PSB1C3+E0240+J23109:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23117:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
Oct 02

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23107:
Measure of the cells' growth and fluorecence on TECAN infiniteM200.
OVERFLOW
PSB1C3+E0240+J23109:
Measure of the cells' growth and fluorecence on TECAN infiniteM200.
TECAN FAILED
PSB1C3+E0240+J23111:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 (17h00-00h00).

PSB1C3+E0240+J23117:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 (9h00-16h00).

Oct 01

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23100:
Measure of the cells' growth and fluorecence on TECAN infiniteM200.
OVERFLOW
PSB1C3+E0240+J23105:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 40-100nM (10h30-17h30).

PSB1C3+E0240+J23107:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23109:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23111:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23117:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
Sep 30

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23100:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23105:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23109:
The pre-culture didn't grow.
Sep 29

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23109:
PCR colony of 8 colonies for each construction following protocol table 1 and 2.

Pre-culture from colony 1 for glycerol stock and TECAN in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23114:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 20-60nM (15h00-22h00).

Sep 28

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23100:
Measure of the cells' growth and fluorecence on TECAN infiniteM200.
OVERFLOW
PSB1C3+E0240+J23108:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 60-20nM (14h00-21h00).

Sep 27

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23100:
Measure of the cells' growth and fluorecence on TECAN infiniteM200.
OVERFLOW
New pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23107:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 40-100nM (10h30-17h30).

PSB1C3+E0240+J23114:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 (9h30-16h30).

PSB1C3+E0240+J23108:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23118:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 5-40nM (18h30-1h30).

Sep 26

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23100:
Measure of the cells' growth and fluorecence on TECAN infiniteM200.
OVERFLOW
New pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23107:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 REPORTED.
New pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23110:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 40-100nM (15h00-22h00).

PSB1C3+E0240+J23112:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 60-20nM (15h00-22h00).

PSB1C3+E0240+J23114:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23118:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
Sep 25

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23100:
Measure of the cells' growth and fluorecence on TECAN infiniteM200.
OVERFLOW
New pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23107:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23110:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23112:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
Sep 24

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23100:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23101:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 40-100nM (19h30-2h30).

PSB1C3+E0240+J23113:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 10-40nM (10h-17h).

PSB1C3+E0240+J23119:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 with rank of GFP 40-80nM (20h40-3h40).

Sep 23

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23101:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23107:
Measure of the cells' growth and fluorecence on TECAN infiniteM200.
OVERFLOW
PSB1C3+E0240+J23113:
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23118:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 (Xh00-Xh00).

PSB1C3+E0240+J23119:
Measure of the cells' growth and fluorecence on TECAN infiniteM200.
TECAN FAILED
Sep 22

Construction n°1: PSB1C3 with I20260
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23100:
- Pre-culture from glycerol in 3mL of LB+Cam (1:1000). Incubation 16h-18h at 37°C.
- Measure of the cells' growth and fluorecence on TECAN infiniteM200 (18h30-2h30).
OVERFLOW
PSB1C3+E0240+J23104:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 (1h30-8h30).

PSB1C3+E0240+J23105:
- Pre-culture from glycerol in 3mL of LB+Cam (1:1000). Incubation 16h-18h at 37°C.
- Measure of the cells' growth and fluorecence on TECAN infiniteM200 (18h30-2h30).

PSB1C3+E0240+J23107:
Pre-culture from glycerol in 3mL of LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23115:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 (10h50-17h50).

PSB1C3+E0240+J23116:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 (11h00-18h00).

PSB1C3+E0240+J23117:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 (1h00-8h00).

PSB1C3+E0240+J23118:
Pre-culture from glycerol in 3mL of LB+Cam (1:1000). Incubation 16h-18h at 37°C.
Sep 19

Construction n°1: PSB1C3 with I20260
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Measure of the cells' growth and fluorecence on TECAN infiniteM200 (00h-07h).

Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23101:

PSB1C3+E0240+J23102:

PSB1C3+E0240+J23104:
Pre-culture from glycerol in 3mL of LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23105:
glycerol stock preparation from LB culture of the 17th September.
PSB1C3+E0240+J23107:
glycerol stock preparation from LB culture of the 17th September.
PSB1C3+E0240+J23110:
Pre-culture does'nt grow. The TECAN measure is reported.
PSB1C3+E0240+J23115:
Pre-culture from glycerol in 3mL of LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23116:
Pre-culture from glycerol in 3mL of LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23117:
Pre-culture from glycerol in 3mL of LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23114:
- PCR colony of 5 colonies for each construction following protocol table 1 and 2.
A pre-culture was made with one colony to make a glycerol stock.
Sep 18

Construction n°1: PSB1C3 with I20260
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23100:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 (11h-16h).
OVERFLOW
PSB1C3+E0240+J23101:
Measure of the cells' growth and fluorecence on TECAN infiniteM200.
TECAN FAILED
PSB1C3+E0240+J23103:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 (16h30-23h30).

PSB1C3+E0240+J23105:
pre-culture of the colony 5 in LB+Cam (1:1000) according to the result of PCR of 13th September. Incubation overnight at 37°C.
PSB1C3+E0240+J23107:
pre-culture of the colony 5 in LB+Cam (1:1000) according to the result of PCR of 13th September. Incubation overnight at 37°C.
PSB1C3+E0240+J23110:
Pre-culture from glycerol in 3mL of LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23115:
Measure of the cells' growth and fluorecence on TECAN infiniteM200 (12h-19h).

PSB1C3+E0240+J23104, PSB1C3+E0240+J23112, PSB1C3+E0240+J23113: PCR colony of 6 colonies for each construction following protocol table 1 and 2.
Migration of the PCR products
PCR purification of clones with the good construction
Sep 17

Construction n°1: PSB1C3 with I20260
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23100:
Pre-culture from glycerol in 3mL of LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23101:
Pre-culture from glycerol in 3mL of LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23102:
- glycerol stock preparation from LB culture from the 14th September.
- Measure of the cells' growth and fluorecence on TECAN infiniteM200.(18h30-1h30):

PSB1C3+E0240+J23106:
- glycerol stock preparation from LB culture from the 14th September.
- Measure of the cells' growth and fluorecence on TECAN infiniteM200.(2h-9h):

PSB1C3+E0240+J23115 - PSB1C3+E0240+J23115:
- PCR colony of 5 colonies for each construction following protocol table 1 and 2.
A pre-culture was made with one colony for each construction to make a glycerol stock.
PSB1C3+E0240+J23117 - PSB1C3+E0240+J23119:
- PCR colony of 6 colonies for each construction following protocol table 1 and 2.
A pre-culture was made with one colony for each construction to make a glycerol stock.
Sep 16

Construction n°1: PSB1C3 with I20260
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Pre-culture from glycerol stock in LB+Cam (1:1000). Incubation 16h-18h at 37°C.
Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23100:
Pre-culture from glycerol in 3mL of LB+Cam (1:1000). Incubation 16h-18h at 37°C.
PSB1C3+E0240+J23106:
Pre-culture from glycerol in 3mL of LB+Cam (1:1000). Incubation 16h-18h at 37°C.
Sep 15

Construction n°1: PSB1C3 with I20260
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23101:
Measure of the cells' growth and fluorecence on TECAN infiniteM200.
TECAN FAILED
PSB1C3+E0240+J23106:
pre-culture of the colony 3 in LB+Cam (1:1000) according to the result of PCR of 13th September. Incubation 16h at 37°C.
Sep 14

Construction n°1: PSB1C3 with I20260
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23101:
pre-culture of the colony 3 in LB+Cam (1:1000). Incubation 16h at 37°C.
PSB1C3+E0240+J23105, PSB1C3+E0240+J23106 and PSB1C3+E0240+J23107:
- PCR colony of 7 colonies for each construction following protocol table 1 and 2.
A pre-culture was made with one colony for each construction to make a glycerol stock.
A pre-culture was made with one colony for each construction to make a glycerol stock.
Sep 13

Construction n°1: PSB1C3 with I20260
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23101 and PSB1C3+E0240+J23118:
glycerol stock preparation from LB culture from the 11th September.
Sep 12

Construction n°1: PSB1C3 with I20260
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23101, PSB1C3+E0240+J23118, PSB1C3+E0240+J23105, PSB1C3+E0240+J23106 and PSB1C3+E0240+J23107:
- Transformation plate from the 10th September observation. There were 50 colonies for each constructions.
PSB1C3+E0240+J23101 and PSB1C3+E0240+J23118,
- PCR colony of 3 colonies for each construction following protocol table 1 and 2.
- PCR purification of sample 1 and 4 of the Gel 1 were purified with the NucleoSpin kit (Macherey Nagel). DNA was quantify by Nanodrop 2000.
- Preparation of samples to sequencing. N° XX
- Preparation of 3 ml cultures LB Cam. Incubation overnight at 37°C.
PSB1C3+E0240+J23100, PSB1C3+E0240+J23102, PSB1C3+E0240+J23103, PSB1C3+E0240+J23104 and PSB1C3+E0240+J23118,
- PCR colony of 1 colony for each construction following protocol table 1 and 2.

GEL 2: 1% agarose gel.
Lane 1: PCR product of 1 colonies of the PSB1C3+E0240+J23100 construction.
Lane 2: PCR product of 1 colonies of the PSB1C3+E0240+J23102 construction.
Lane 3: PCR product of 1 colonies of the PSB1C3+E0240+J23103 construction.
Lane 4: PCR product of 1 colonies of the PSB1C3+E0240+J23104 construction.
A pre-culture was made for each colony to make a glycerol stock.
Sep 11

Construction n°1: PSB1C3 with I20260
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Other constructions of the Anderson library of constitutive promoters
Miniprep disgestion of PSB1C3+E0240 plasmid.
- 1 µl EcoRI
- 1 µl XbaI
- 2 µl buffer 2.1
- 0.2 µl BSA
- 5 µl template (miniprep)
- 10.8 µl miliQ watter
Miniprep disgestion of BBa_J61002+J23101, BBa_J61002+J23118, BBa_J61002+J23105, BBa_J61002+J23106 and BBa_J61002+J23107 plasmids.
- 1 µl EcoRI
- 1 µl SpeI
- 2 µl buffer 2.1
- 0.2 µl BSA
- 5 µl template (miniprep)
- 10.8 µl miliQ watter
Ligation to obtain constructions PSB1C3+E0240+J23101, PSB1C3+E0240+J23118, PSB1C3+E0240+J23105, PSB1C3+E0240+J23106 and PSB1C3+E0240+J23107.
- 1 µl T4 DNA ligase
- 6 µl Insert (digested BBa_J61002+J23101, BBa_J61002+J23118, BBa_J61002+J23105, BBa_J61002+J23106 and BBa_J61002+J23107)
- 2 µl vector (digested PSB1C3+E0240)
- 2 µl buffer
- 9 µl miliQ watter
Transformation in E. coli DH5 alpha with the protocol of the 15th of August. Bacteria were plated on Chloramphenicol agar plates and incubated at 37°C overnight.
Sep 10

Construction n°1: PSB1C3 with I20260
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Other constructions of the Anderson library of constitutive promoters
Sep 09

Construction n°1: PSB1C3 with I20260
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Other constructions of the Anderson library of constitutive promoters
Sep 07

Construction n°1: PSB1C3 with I20260
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Other constructions of the Anderson library of constitutive promoters
PSB1C3+E0240+J23111:
PCR colony of 7 colonies for each construction following protocol table 1 and 2.

Pre-culture of colony 2 for grycerol stock
Sep 06

Construction n°1: PSB1C3 with I20260
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Other constructions of the Anderson library of constitutive promoters
Sep 05

Construction n°1: PSB1C3 with I20260
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Other constructions of the Anderson library of constitutive promoters
Sep 04

Construction n°1: PSB1C3 with I20260
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Other constructions of the Anderson library of constitutive promoters
Sep 03

Construction n°1: PSB1C3 with I20260
Analyse of sequencing results. The two constructions were ok although the colony 4 had a better match. So we decided to keep this colony to perform promoter activity assays.
26DJ96
GTCAGTGAGCGAGGNAAGCCTGCATAACGCGAAGTAATCTTTTCGGTTTTAAAGAAAAAGGGCAGGGTGGTGACACCTTGCCCTTTTTTGCCGGACTGCAGC GGCCGCTACTAGTATATAAACGCAGAAAGGCCCACCCGAAGGTGAGCCAGTGTGACTCTAGTAGAGAGCGTTCACCGACAAACAACAGATAAAACGAAAGGCCCAGT CTTTCGACTGAGCCTTTCGTTTTATTTGATGCCTGGCTCTAGTATTATTATTTGTATAGTTCATCCATGCCATGTGTAATCCCAGCAGCTGTTACAAACTCAAGAAG GACCATGTGGTCTCTCTTTTCGTTGGGATCTTTCGAAAGGGCAGATTGTGTGGACAGGTAATGGTTGTCTGGTAAAAGGACAGGGCCATCGCCAATTGGAGTATTTT GTTGATAATGGTCTGCTAGTTGAACGCTTCCATCTTCAATGTTGTGTCTAATTTTGAAGTTAACTTTGATTCCATTCTTTTGTTTGTCTGCCATGATGTATACATTG TGTGAGTTATAGTTGTATTCCAATTTGTGTCCAAGAATGTTTCCATCTTCTTTAAAATCAATACCTTTTAACTCGATTCTATTAACAAGGGTATCACCTTCAAACTT GACTTCAGCACGTGTCTTGTAGTTCCCGTCATCTTTGAAAAATATAGTTCTTTCCTGTACATAACCTTCGGGCATGGCACTCTTGAAAAAGTCATGCTGTTTCATAT GATCTGGGTATCTCGCANAGCATTGAACACCATAACCGAAAGTAGTGACAAGTGTTGGCCATGGAACAGGTAGTTTTCCAGTAGTGCAAATAAATTTAAGGGTAAGT TTTCCGTATGTTGCATCACCTTCACCCTCTCCACTGACAGAAAATTTGTGCCCATTAACATCACCATCTAATTCAACAAGAATTGGGACAACTCCAGTGAAAAGTTC TTCTCCTTTACGCATCTAGTACTTTCCTNNGTGACTCTAGTAGCTANCCATAANNCCTAGGAACTGANCTAGCTG
26DJ97
GATTACTANNAATAGGCGTATCACGAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTAAGGATGATTTCTGGAATTCGCGGCCGCTTCTAGAGTT TACAGCTAGCTCAGTCCTAGGTATTATGCTAGCTACTAGAGTCACACAGGAAAGTACTAGATGCGTAAAGGAGAAGAACTTTTCACTGGAGTTGTCCCAATTCTTGT TGAATTAGATGGTGATGTTAATGGGCACAAATTTTCTGTCAGTGGAGAGGGTGAAGGTGATGCAACATACGGAAAACTTACCCTTAAATTTATTTGCACTACTGGAA AACTACCTGTTCCATGGCCAACACTTGTCACTACTTTCGGTTATGGTGTTCAATGCTTTGCGAGATACCCAGATCATATGAAACAGCATGACTTTTTCAAGAGTGCC ATGCCCGAAGGTTATGTACAGGAAAGAACTATATTTTTCAAAGATGACGGGAACTACAAGACACGTGCTGAAGTCAAGTTTGAAGGTGATACCCTTGTTAATAGAAT CGAGTTAAAAGGTATTGATTTTAAAGAAGATGGAAACATTCTTGGACACAAATTGGAATACAACTATAACTCACACAATGTATACATCATGGCAGACAAACAAAAGA ATGGAATCAAAGTTAACTTCAAAATTAGACACAACATTGAAGATGGAAGCGTTCAACTAGCAGACCATTATCAACAAAATACTCCAATTGGCGATGGCCCTGTCCTT TTACCAGACAACCATTACCTGTCCACACAATCTGCCCTTTCGAAAGATCCCAACGAAAAGAGAGACCACATGGTCCTTCTTGAGTTTGTAACAGCTGCTGGGATTAC ACATGGCATGGATGAACTATACAAATAATAATACTAGAGCCAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCG GTGAACGCTCTCTACTAGAGTCACACTGGCTCACCTTCGGGTGGGCCTTTCTGCGTTTATATACTAGTAGCGGCCGCTGCAGTCCGGCAAANAAGGGCAAGGTGTCA CCACCCTGCCCTTTTTCTTTAAAACCGAAAANATTACTT
26DJ98
AGTCAGTGAGCGAGGAAGCCTGCATAACGCGAAGTAATCTTTTCGGTTTTAAAGAAAAAGGGCAGGGTGGTGACACCTTGCCCTTTTTTGCCGGACTGCAGC GGCCGCTACTAGTATATAAACGCAGAAAGGCCCACCCGAAGGTGAGCCAGTGTGACTCTAGTAGAGAGCGTTCACCGACAAACAACAGATAAAACGAAAGGCCCAGT CTTTCGACTGAGCCTTTCGTTTTATTTGATGCCTGGCTCTAGTATTATTATTTGTATAGTTCATCCATGCCATGTGTAATCCCAGCAGCTGTTACAAACTCAAGAAG GACCATGTGGTCTCTCTTTTCGTTGGGATCTTTCGAAAGGGCAGATTGTGTGGACAGGTAATGGTTGTCTGGTAAAAGGACAGGGCCATCGCCAATTGGAGTATTTT GTTGATAATGGTCTGCTAGTTGAACGCTTCCATCTTCAATGTTGTGTCTAATTTTGAAGTTAACTTTGATTCCATTCTTTTGTTTGTCTGCCATGATGTATACATTG TGTGAGTTATAGTTGTATTCCAATTTGTGTCCAAGAATGTTTCCATCTTCTTTAAAATCAATACCTTTTAACTCGATTCTATTAACAAGGGTATCACCTTCAAACTT GACTTCAGCACGTGTCTTGTAGTTCCCGTCATCTTTGAAAAATATAGTTCTTTCCTGTACATAACCTTCGGGCATGGCACTCTTGAAAAAGTCATGCTGTTTCATAT GATCTGGGTATCTCGCANAGCATTGAACACCATAACCGAAAGTAGTGACAAGTGTTGGCCATGGAACAGGTAGTTTTCCAGTAGTGCAAATAAATTTAAGGGTAAGT TTTCCGTATGTTGCATCACCTTCACCCTCTCCACTGACAGAAAATTTGTGCCCATTAACATCACCATCTAATTCAACAAGAATTGGGACAACTCCAGTGAAAAGTTC TTCTCCTTTACGCATCTAGTACTTTCCTGTGTGACTCTAGTAGCTAGCATAATACCTAGGACTGAGCTNNNTGTAAACTCTNNNANCGGCC
26DJ99
TAGGCGTATNANGAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTAAGGATGATTTCTGGAATTCGCGGCCGCTTCTAGAGTTTACAGCTAGCTC AGTCCTAGGTATTATGCTAGCTACTAGAGTCACACAGGAAAGTACTAGATGCGTAAAGGAGAAGAACTTTTCACTGGAGTTGTCCCAATTCTTGTTGAATTAGATGGT GATGTTAATGGGCACAAATTTTCTGTCAGTGGAGAGGGTGAAGGTGATGCAACATACGGAAAACTTACCCTTAAATTTATTTGCACTACTGGAAAACTACCTGTTCCA TGGCCAACACTTGTCACTACTTTCGGTTATGGTGTTCAATGCTTTGCGAGATACCCAGATCATATGAAACAGCATGACTTTTTCAAGAGTGCCATGCCCGAAGGTTAT GTACAGGAAAGAACTATATTTTTCAAAGATGACGGGAACTACAAGACACGTGCTGAAGTCAAGTTTGAAGGTGATACCCTTGTTAATAGAATCGAGTTAAAAGGTATT GATTTTAAAGAAGATGGAAACATTCTTGGACACAAATTGGAATACAACTATAACTCACACAATGTATACATCATGGCAGACAAACAAAAGAATGGAATCAAAGTTAAC TTCAAAATTAGACACAACATTGAAGATGGAAGCGTTCAACTAGCAGACCATTATCAACAAAATACTCCAATTGGCGATGGCCCTGTCCTTTTACCAGACAACCATTAC CTGTCCACACAATCTGCCCTTTCGAAAGATCCCAACGAANAGAGAGACCACATGGTCCTTCTTGAGTTTGTAACAGCTGCTGGGATTACACATGGCATGGATGAACTA TACAAATAATAATACTAGAGCCAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCGGTGAACGCTCTCTACTAGAG TCACACTGGCTCACCTTCGGGTGGGCCTTTCTGCGTTTATATACTAGTAGCGGCCGCTGCAGTCCGGCAAAAAAGGGCAAGGTGTCACCACCCTGCCCTTTTTCTTTA AAACCGAAAANATTACTTCNCGTTATGCAGGCTTCCTCGCTCACTGACTCGCTGCGCTCGGTCGTTCGGCTGCGGCGNACGG
26EA00
Impossible to read
26EA01
Impossible to read
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Other constructions of the Anderson library of constitutive promoters
Miniprep cultures from the 2cnd of september was purified with the NucleoSpin Plasmid kit (Macherey-Nagel). DNA was quantify with the Nanodrop 2000.
Glycerol stock were prepared started from the 16 cultures from the 2cnd of september (750 µl of culture + 750 µL of 10% glycerol) and stored at -80°C. Sep 02

Other constructions of the Anderson library of constitutive promoters
PCR colony were performed on colonies from transformations of the 29th August. Used protocol was the same as Table 1 and 2 with the Q5 high fidelity enzyme.
PCR product of first colony of each transformation was loaded on 1% agarose gel and gel run 45 minutes at 110 mV. 3 ml LB culture were started from tested colonies which presented the right PCR product profile. Cultures were incubated overnight at 37°C. Sep 01

Other constructions of the Anderson library of constitutive promoters
Aug 31

Other constructions of the Anderson library of constitutive promoters
Transformation plates observation. PCR colony were performed on 8 colonies per transformation plate. PCR products were loaded on 1% agarose gel and gel ran one 45 minutes. There were nothing on gel, reason is probably a mix and temperature problem.
Aug 30

Construction n°1: PSB1C3 with I20260
Selected colonies PCR products from the 28/08/2014 were purified with the NucleoSpin kit (Macherey Nagel). DNA was quantify by Nanodrop 2000.
- Colony n°2: 30.7 ng/µl and A260/280 1.92
- Colony n°4: 52.3 ng/µl and A260/280 1.75
- Colony n°7: 26.1 ng/µL and A260/280 1.72
Samples were send to sequencing. N° 26DJ96, 26DJ97, 26DJ98, 26DJ99, 26EA00 and 26EA01.
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: PSB1C3 with K823012-E1010
Selected colonies PCR products from the 28/08/2014 were purified with the NucleoSpin kit (Macherey Nagel). DNA was quantify by Nanodrop 2000.
- Colony n°2: 41.4 ng/µl
- Colony n°3: 65.8 ng/µl
- Colony n°4: 27.4 ng/µL
Colony n°3 was sent to sequence with the identification 26AE07 (VF2) and 26AE08 (VR)
Other constructions of the Anderson library of constitutive promoters
Transformation in DH5 alpha chemocompetent E. coli as doing on 21th August, with ampicilline LB agar plates
Incubation overnight at 37°C.
Aug 29

Contruction n°2: PSB1C3 with J23101-E1010
Contruction n°3: PSB1C3 with K823012-E1010
PCR colony was done on 7 colonies:
- Steril H2O:34,5µL
- 5X Q5 Reaction buffer: 10µL
- Template:2µL
- 10mM dNTP: 1µL
- 10µM Primer forward VF2: 1µL
- 10µM Primer reverse VR: 1µL
- Q5 DNA Polymerase: 0,5µL
A migration of PCR product was done. We expect to observe in each wells a band of 1176bp:

6 colonies have a band at 1200 bp around so we decided to choose colonies n°2, 3 and 4.
Other constructions of the Anderson library of constitutive promoters
Promoters were resuspended from 2014 distribution kit plates with 10 µl of sterile MiliQ watter and stored at -20°C.
Aug 28

Contruction n°1: pSB1C3 with I20260
Contruction n°2: pSB1C3 with J23101-E1010
Contruction n°3: pSB1C3 with K823012-E1010
At the end of the day, there were 3 colonies that have grown
Aug 27

Construction n°1: PSB1C3 with I20260
Loading of the PSB1C3 digestion by EcoRI and PstI from the 25th August, on a 1% agarose gel. The 40 µl were loaded with 8 µl of loading dye 6X. Migration was performed 1 hour at 110 mV.
Construction n°2: PSB1C3 with J23101-E1010
Construction n°3: pSB1C3 with K823012-E1010
Digestion of the insert pSB1C3 (J23115) with EcoRI and XbaI:
- Steril H2O: 10,8µL
- Buffer 2.1 NEB : 2µL
- BSA: 0,2µL
- DNA: 5µL
- EcoRI-HF: 1µL
- XbaI: 1µl
Digestion of the vector pSB1C3(E0240) with EcoRI and SpeI:
- Steril H2O: 10,8µL
- Buffer 2.1 NEB : 2µL
- BSA: 0,2µL
- DNA: 5µL
- EcoRI-HF: 1µL
- SpeI: 1µl
Mix were incubated at 37°C during 45mn then at 80°C during 20 mn
Ligation:
- Steril H2O: 9µL
- Insert (J23115):6µL
- Vector (E0240): 2µL
- 10X T4 DNA ligase reaction buffer: 2µL
Mix was incubated at 10mn at room temperature then at 80°C during 20mn before the transformation in DH5a chimiocompetent with 3µL of the ligation product with the same protocol
Aug 26

Aug 25

Aug 24

Contruction n°1: PSB1C3 with I20260
Digestion of PSB1C3 and PSB1K3-I20260 by EcoRI and PstI
- 1 µl EcoRI-HF
- 1 µl PstI
- 2 µl buffer
- 0.2 µl BSA
- 13.7 or 76 µl respectively for I20260 and PSB1C3
- 10.8 µl miliQ watter
Incubation 45 minutes at 37°C. Denaturation step 20 minutes at 80 °C.
Loading on a 1% agarose gel to perform an extraction from gel
Contruction n°2: PSB1C3 with J23101-E1010
Aug 23

Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
Transformation plate observation:
- BBa_J23101: 11 isolated colonies
- BBa_K823012: 100-150 isolated colonies
A PCR was performed with 8 colonies for BBa_K823012 and for BBa_J23101 following the protocol Table 3 and 4.
To verify PCR products, 10 µl was loaded on a 1% agarose gel with 2 µl of loading dye 6X.
We expected to obtain a band between 300 and 400 bp. So we decided to amplify the colony n°8. The colony was seed in 5 ml of LB medium and 5 µl of chloramphenicol. Culture was incubated overnight at 37°C with 200 rpm agitation.
We expected to obtain a band around 1200 bp. So we decided to amplify the colony n°2 corresponding to lane 11. The colony was seed in 5 ml of LB medium and 5 µl of chloramphenicol. Culture was incubated overnight at 37°C with 200 rpm agitation. Aug 22

Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
Transformations of registry parts (BBa_K823012 and J23101) were performed to obtain colonies with plasmid containing parts for future amplifications.
The transformation was performed on DH5 alpha E. coli, as followed:
- Remove E. coli competent tubes from -80°C and keep it on ice
- Add 3 µl of template (here solubilized plasmids from the registry distribution kit) and mix gently
- Incubate 10 minutes on ice
- Perform an heat shock 30 seconds at 42°C
- Incubate 2 minutes on ice
- Add 2 ml of LB medium and incubate 60 minutes at 37°C with an agitation at 200 rpm
- Plate 200 µl of BBa_K823012 or BBa_J23101 on a chloramphenicol LB agar plate.
- Incubate plate overnight at 37°C

Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
Miniprep cultures from the 19th August was purified with the NucleoSpin Plasmid kit (Macherey-Nagel). DNA was quantify with the Nanodrop 2000. DNA concentrations and A260/280 were respectively for BBa_E0240 and BBa_I20260: 42 ng/µl and 1.91 ; 13.7 ng/µl and 1.71.
A glycerol stock was prepared for each culture and stored at -80°C.

Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
Preparation of a 1% agarose gel: 1.01 g of Top Vision agarose (Thermo Scientific) + 100 ml of TAE 1X. Microwave 30s by 30s until agarose total dissolution Gel was cooling down until to be lukewarm, one BET drop was added. Gel was loaded with 10µl per sample previously added with 2 µl of loading dye 6X, and 5 µl for ladders. Gel running 45 minutes at 100 mV in TAE 1X buffer.
Purified PCR product aliquot was send to sequencing. The sequencing number were 26DJ68, 69, 70, 71.
Preparation of miniprep culture of BBa_I20260 and BBa_E0240 selected colonies in 5 ml of LB with respectively 5 µl of Kana or chloramphenicol solution stock. Incubation overnight at 37°C and 300 rpm. Aug 19

Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
Sequencing results were received for BBa_J23101 (26DJ62 and 26DJ63) and BBa_K823012(26DJ64 and 26DJ65) PCR products.
26DJ62
TCAGANNAAAAAAATCCTTAGCTNNCGCTAAGGATGANNTCTGGAATTCGCGGCCGCTTCTAGAGTTTACAGCTAGCTCAGTCCTAGGTATTATGCTAGCTACTAGTAG CGGCCGCTGCAGTCCGGCAAAAAAGGGCAAGGTGTCACCACCCTGCCCTTTTTCTTTAAAACCGAAAAGATTACTTCGCGTTATGCAGGCTTCCTCGCTCACTGACTC GCTGCGCTCGGTCGTTCGGCTGCGGCGAGCGGTATCAGCTCACTCAGAG
26DJ63
GAGCGCANCGAGTCAGTGAGCGAGGAAGCCTGCATAACGCGAAGTAATCTTTTCGGTTTTAAAGAAAAAGGGCAGGGTGGTGACACCTTGCCCTTTTTTGCCGGACTG CAGCGGCCGCTACTAGTAGCTAGCATAATACCTAGGACTGAGCTAGCTGTAAACTCTAGAAGCGGCCGCGAATTCCAGAAATCATCCTTAGCGAAAGCTAAGGATTTTT TTTATCTGAAATTCTGCCTCGTGATACGCCTATTTTTATAGGTTAATGTCATGATAATAATGGTTTCTTAGACGCAAGGTGGCAAA
26DJ64
TCAGANNAAAAAAATCCTTAGCTNNCGCTAAGGATGATTTCTGGAATTCGCGGCCGCTTCTAGAGGTTTATAGCTAGCTCATCCTAGGTACAATGCTAGCTACTAG TAGCGGCCGCTGCAGTCCGGCAAAAAAGGGCAAGGTGTCACCACCCTGCCCTTTTTCTTTAAAACCGAAAAGATTACTTCGCGTTATGCAGGCTTCCTCGCTCACTGAC TCGCTGCGCTCGGTCGTTCGGCTGCGGCGAGCGGTATCAGCTCACTCAGG
26DJ65
CNGCGAGTCAGTNAGCGAGGAAGCCTGCANNACGCGAAGTNATCTTTTTCGGTTTTAAAGAAAAAGGGCAGGGTGGTGACACCTNGCCCTTTTTTGCCGGACTGCAGC GGCCGCTACTAGTAGCTAGCATTGTACCTAGGACTGAGCTAGCTATAAACTCTAGAAGCGGCCGCGAATTCCAGAAATCATCCTTAGCGAAAGCTAAGGATTTTTTTTA TCTGAAATTCTGCCTCGTGATACGCCTATTTTTATAGGTTAATGTCATGATAATAATGGTTTCTTAAC
The mapping was perfect for BBa_J23101 for BBa_K823012.
Sequencing results were received for BBa_E0240 (26DJ68 and 26DJ69) and BBa_I20260 (26DJ70 and 26DJ71) PCR products.
26DJ68
GAGCNCAGCGAGTCAGTGAGCGAGGAAGCCTGCATAACGCGAAGTAATCTTTTCGGTTTTAAAGAAAAAGGGCAGGGTGGTGACACCTTGCCCTTTTTTGCCGGACTG CAGCGGCCGCTACTAGTATATAAACGCAGAAAGGCCCACCCGAAGGTGAGCCAGTGTGACTCTAGTAGAGAGCGTTCACCGACAAACAACAGATAAAACGAAAGGCCC AGTCTTTCGACTGAGCCTTTCGTTTTATTTGATGCCTGGCTCTAGTATTATTATTTGTATAGTTCATCCATGCCATGTGTAATCCCAGCAGCTGTTACAAACTCAAGA AGGACCATGTGGTCTCTCTTTTCGTTGGGATCTTTCGAAAGGGCAGATTGTGTGGACAGGTAATGGTTGTCTGGTAAAAGGACAGGGCCATCGCCAATTGGAGTATTT TGTTGATAATGGTCTGCTAGTTGAACGCTTCCATCTTCAATTTGTGTCTAATTTTGAAGTTAACTTTGATTCCATTCTTTTGTTTGTCTGCCATGATGTATACATTGT GTGAGTTATAGTTGTATTCCAATTTGTGTCCAAGAATGTTTCCATCTTCTTTAAAATCAATACCTTTTAACTCGATTCTATTAACAAGGGTATCACCTTCAAACTTGA CTTCAGCACGTGTCTTGTAGTTCCCGTCATCTTTGAAAAATATAGTTCTTTCCTGTACATAACCTTCGGGCATGGCACTCTTGAAAAAGTCATGCTGTTTCATATGAT CTGGGTATCTCGCAAAGCATTGAACACCATAACCGAAAGTAGTGACAAGTGTTGGCCATGGAACAGGTAGTTTTCCAGTAGTGCAAATAAATTTAAGGGTAAGTTTTC CGTATGTTGCATCACCTTCACCCTCTCCACTGACAGAAAATTTGTGCCCATTAACATCACCATCTAATTCAACAAGAATTGGGACAACTCCAGTGAANAGTTCTTCTC CTTTACGCATCTAGTACTTTCCTGTGTGACTCTAGTAGCTAGCATAATACCTAGGACTGAGCTAGCTGTNAACTCTANAAGCGGCCGCGAATTCCAGAAATCATCCTT ANCNNAAGCTAAGGATTTTTTTTATCTGNAATTCTGCCTC
26DJ69
GATTACTATNAAATAGGCGTANNANGAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTAAGGATGATTTCTGGAATTCGCGGCCGCTTCTAGAGTTTACAG CTAGCTCAGTCCTAGGTATTATGCTAGCTACTAGAGTCACACAGGAAAGTACTAGATGCGTAAAGGAGAAGAACTTTTCACTGGAGTTGTCCCAATTCTTGTTGAATT AGATGGTGATGTTAATGGGCACAAATTTTCTGTCAGTGGAGAGGGTGAAGGTGATGCAACATACGGAAAACTTACCCTTAAATTTATTTGCACTACTGGAAAACTACC TGTTCCATGGCCAACACTTGTCACTACTTTCGGTTATGGTGTTCAATGCTTTGCGAGATACCCAGATCATATGAAACAGCATGACTTTTTCAAGAGTGCCATGCCCGA AGGTTATGTACAGGAAAGAACTATATTTTTCAAAGATGACGGGAACTACAAGACACGTGCTGAAGTCAAGTTTGAAGGTGATACCCTTGTTAATAGAATCGAGTTAAA AGGTATTGATTTTAAAGAAGATGGAAACATTCTTGGACACAAATTGGAATACAACTATAACTCACACAATGTATACATCATGGCAGACAAACAAAAGAATGGAATCAA AGTTAACTTCAAAATTAGACACAACATTGAAGATGGAAGCGTTCAACTAGCAGACCATTATCAACAAAATACTCCAATTGGCGATGGCCCTGTCCTTTTACCAGACAA CCATTACCTGTCCACACAATCTGCCCTTTCGAAAGATCCCAACGAANAGAGAGACCACATGGTCCTTCTTGAGTTTGTAACAGCTGCTGGGATTACACATGGCATGGA TGAACTATACAAATAATAATACTAGAGCCAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCGGTGAACGCTCTCT ACTAGAGTCACACTGGCTCACCTTCGGGTGGGCCTTTCTGCGTTTATATACTAGTAGCGGCCGCTGCAGTCCGGCNAAAAAGGGCAAGGTGTCACCACCNTGCCCTTT TTCTTTAAAACCGNAAA
26DJ70
AGCGAGTCAGTGAGCGAGGAAGCCTGCATAACGCGAAGTAATCTTTTCGGTTTTAAAGAAAAAGGGCAGGGTGGTGACACCTTGCCCTTTTTTGCCGGACTGCAGCGG CCGCTACTAGTATATAAACGCAGAAAGGCCCACCCGAAGGTGAGCCAGTGTGACTCTAGTAGAGAGCGTTCACCGACAAACAACAGATAAAACGAAAGGCCCAGTCTT TCGACTGAGCCTTTCGTTTTATTTGATGCCTGGCTCTAGTATTATTATTTGTATAGTTCATCCATGCCATGTGTAATCCCAGCAGCTGTTACAAACTCAAGAAGGACC ATGTGGTCTCTCTTTTCGTTGGGATCTTTCGAAAGGGCAGATTGTGTGGACAGGTAATGGTTGTCTGGTAAAAGGACAGGGCCATCGCCAATTGGAGTATTTTGTTGA TAATGGTCTGCTAGTTGAACGCTTCCATCTTCAATGTTGTGTCTAATTTTGAAGTTAACTTTGATTCCATTCTTTTGTTTGTCTGCCATGATGTATACATTGTGTGAG TTATAGTTGTATTCCAATTTGTGTCCAAGAATGTTTCCATCTTCTTTAAAATCAATACCTTTTAACTCGATTCTATTAACAAGGGTATCACCTTCAAACTTGACTTCA GCACGTGTCTTGTAGTTCCCGTCATCTTTGAAAAATATAGTTCTTTCCTGTACATAACCTTCGGGCATGGCACTCTTGAAAAAGTCATGCTGTTTCATATGATCTGGG TATCTCGCAAAGCATTGAACACCATAACCGAAAGTAGTGACAAGTGTTGGCCATGGAACAGGTAGTTTTCCAGTAGTGCAAATAAATTTAAGGGTAAGTTTTCCGTAT GTTGCATCACCTTCACCCTCTCCACTGACAGAAAATTTGTGCCCATTAACATCACCATCTAATTCAACAAGAATTGGGACAACTCCAGTGAAAAGTTCTTCTCCTTTA CGCATCTAGTACTTTCCTGTGTGACTCTAGAAGCGGCCGCGAATTCCAGAAATCATCCTTAGCGATGAATTCCAGAAATCATCCTTANCGAAAGCTNAGGATTTTTTT TATCTGNAATTCTGNCTCG
26DJ71
GATTAAACTATNAATAGGCGTATCACGAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTAAGGATGATTTCTGGAATTCATCGCTAAGGATGATTTCTGGA ATTCGCGGCCGCTTCTAGAGTCACACAGGAAAGTACTAGATGCGTAAAGGAGAAGAACTTTTCACTGGAGTTGTCCCAATTCTTGTTGAATTAGATGGTGATGTTAAT GGGCACAAATTTTCTGTCAGTGGAGAGGGTGAAGGTGATGCAACATACGGAAAACTTACCCTTAAATTTATTTGCACTACTGGAAAACTACCTGTTCCATGGCCAACA CTTGTCACTACTTTCGGTTATGGTGTTCAATGCTTTGCGAGATACCCAGATCATATGAAACAGCATGACTTTTTCAAGAGTGCCATGCCCGAAGGTTATGTACAGGAA AGAACTATATTTTTCAAAGATGACGGGAACTACAAGACACGTGCTGAAGTCAAGTTTGAAGGTGATACCCTTGTTAATAGAATCGAGTTAAAAGGTATTGATTTTAAA GAAGATGGAAACATTCTTGGACACAAATTGGAATACAACTATAACTCACACAATGTATACATCATGGCAGACAAACAAAAGAATGGAATCAAAGTTAACTTCAAAATT AGACACAACATTGAAGATGGAAGCGTTCAACTAGCAGACCATTATCAACAAAATACTCCAATTGGCGATGGCCCTGTCCTTTTACCAGACAACCATTACCTGTCCACA CAATCTGCCCTTTCGAAAGATCCCAACGAANAGAGAGACCACATGGTCCTTCTTGAGTTTGTAACAGCTGCTGGGATTACACATGGCATGGATGAACTATACAAATAA TAATACTAGAGCCAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCGGTGAACGCTCTCTACTAGAGTCACACTGG CTCACCTTCGGGTGGGCCTTTCTGCGTTTATATACTAGTAGCGGCCGCTGCAGTCCGGCAAANAAGGGCAAGGTGTCACCACCNTGCCCTTTTTCTTTAAAACCGNAA AGATTACTTCNNNGTTATGCAGGCTTCCTCGCTCACTGACTCGCTGCGCTCGGTCGTTCGGCT
The mapping were correct for BBa_E0240 and BBa_I20260.
Colony PCRs were performed on 8 colonies for BBa_E0240 and BBa_I20260, form plates from the transformation of 16th August. 49 µl of mix were distributed in each PCR tubes. The One Taq PCR program was applied. Aug 18

Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
Colonies had grown on plates as followed:
- LB agar: more than 1000 colonies for the two biobricks
- LB agar kanamycin: 10 colonies for BBa_E0240 and >40 colonies for BBa_I20260
- LB agar chloramphenicol: >50 colonies for BBa_E0240 and 3 colonies for BBa_I20260
Plate storage at 4°C.
Aug 17
Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
Nothing had grown during the night on both plates. Taking into consideration different hypothesis, the transformation was repeated with a change on step 6 and 7. At step 6, only 1 ml of LB medium was added. To plate, 200 µl of BBa_E0240 and BBa_I20260 were plated on 3 plates: LB agar, LB agar kanamycin and LB agar chloramphenicol.
Aug 16
Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
BBa_E0240 and BBa_I20260 are respectively into a PSB1C3 and a PSB1K3 vector. In order to select colonies, LB complemented with kanamycin or chloramphenicol are necessary. 200 ml of LB agar medium was prepared with 7 g of LB Agar powder(Sigma) into 200 ml of miliQ water. After complete dissolution, the solution was sterilized in the Tuttmauer 2540ML apparatus, STE 20 minutes at 121°C and EXT+DRY 15 minutes. 50 µl of kanamycin stock solution (25 mg/L) was added to 50 ml of LB agar to pour 2 plates. 150 µl of chloramphenicol stock solution was added in the 150 LB agar remaining ml to pour 6 plates.
The transformation was performed into DH5 alpha ''E. coli'', as followed:
- Remove E. coli competent tubes from -80°C and keep it on ice
- Add 1 µl of template (here solubilized plasmids from the registry distribution kit) and mix gently
- Incubate 10 minutes on ice
- Perform an heat shock 30 seconds at 42°C
- Incubate 2 minutes on ice
- Add 3 ml of LB medium and incubate 60 minutes at 37°C with an agitation at 200 rpm
- Plate 200 µl of BBa_E0240 on a chloramphenicol LB agar plate and BBa_I20260 on a Knamycin LB agar plate
- Incubate plate overnight at 37°C

Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
To amplify BBa_I20260 and BBa_0240, a PCR was perform with the mix described Table 1. The program IGEM Q5 PCR was applied, see Table 2. PCR products were cleaned with the GeneJET purification kit (Thermo Scientific)followed by DNA quantification with the NanoDrop 2000 (Thermo Scientific). Purified samples of I20260 and E1010 had respectively following concentration 30.9 ng/µl and 43.3 ng/µl.
Preparation of a 1% agarose gel: 0.52 g of Top Vision agarose (Thermo Scientific) + 50 ml of TAE 1X. Microwave 30s by 30s until agarose total dissolution Gel was cooling down until to be lukewarm, one BET drop was added. Gel was loaded with 20 µl of BBa_E0240 PCR product and BBa_I20260 purified PCR product added with 4 µl of loading dye, at 10µl of the mix per well.
Gel running 45 minutes at 100 mV in TAE 1X buffer.
3 pieces of gel were sampled in four tubes to perform a DNA purification from gel. The DNA purification was made with the GeneJET purification kit (Thermo Scientific).
A verification electrophoresis was performed on a 1% agarose gel.
The major part of the DNA was lost during the purification because the amount of was to weak to be purified from gel. So we decide do transform ''E. coli'' to isolate a colony containing the desired plasmid for BBa_E0240 and BBa_I20260.
Aug 14
Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
PCR products were cleaned with the GeneJET purification kit (Thermo Scientific)followed by DNA quantification with the NanoDrop 2000 (Thermo Scientific).
Purified PCR product of BBa_J61002/J23101 and BBa_J61002/K823012 concentration were respectively: 53.9 ng/µl and 54.1 ng/µl.
Preparation of a 1% agarose gel: 0.56 g of Top Vision agarose (Thermo Scientific) + 50 ml of TAE 1X. Microwave 30s by 30s until agarose total dissolution Gel was cooling down until to be lukewarm, one BET drop was added. Gel was loaded with 10µl per sample previously added with 2 µl of loading dye 6X, and 5 µl for ladders. Gel running 45 minutes at 100 mV in TAE 1X buffer.
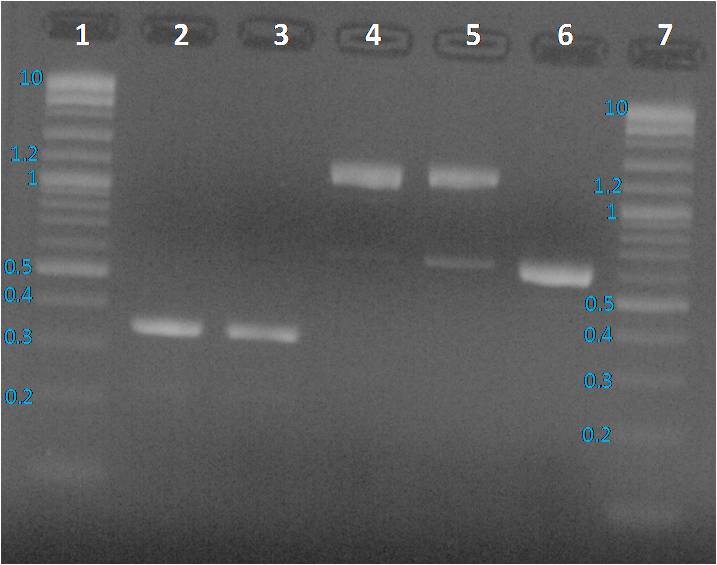
Profiles were same as before PCR clean up. To sequence the amplified parts, we decided to a purification of fragments from the gel as presented below.
Preparation of a 1% agarose gel: 0.56 g of Top Vision agarose (Thermo Scientific) + 50 ml of TAE 1X. Microwave 30s by 30s until agarose total dissolution. Gel was cooling down until to be lukewarm, one BET drop was added. Gel was loaded with 30 µl of BBa_E0240 purified PCR product and BBa_I20260 purified PCR product added with 6 µl of loading dye, at 10µl of the mix per well. Gel running 45 minutes at 100 mV in TAE 1X buffer.
Four pieces of gel were sampled in four tubes to perform a DNA purification from gel.
Aug 13

Contructions PSB1C3 with I20260, J23101-E1010 and K823012-E1010
The 4 needed parts are: BBa_J23101, BBa_K823012, BBa_E0240 and BBa_I20260. Corresponding wells were located on 2014 Distribution kit plates and resuspended with 10 µL steril water. That permits to obtain a DNA concentration around 0.2 ng/µl (according to the registry. Solutions were transferred into 1 ml eppendorf tubes and stored at -20°C.
To amplify fragments, a PCR was performed on the 4 constructions, with the mix described on table 1.
Distribution of 49 µl of mix per PCR tube. Application of program IGEM Q5 PCR.
Preparation of a 1% agarose gel: 0.56 g of Top Vision agarose (Thermo Scientific) + 50 ml of TAE 1X.Microwave 30s by 30s until agarose total dissolution. Gel was cooling down until to be lukewarm, one BET drop was added. Gel was loaded with 10µl per sample previously added with 2 µl of loading dye 6X, and 5 µl for ladders. Gel running 45 minutes at 100 mV in TAE 1X buffer.
We expected to obtain one band per PCR sample corresponding to the interesting amplified fragment. For Lane 2, 3 and 6 it was ok. We had the expected profile. By contrast 2 bands were visible, one at the expected size (around 1200 bp) and another around 600 bp. We decided to perform a purification on gel of each bands.
Aug 12
 "
"
































