Team:Oxford/codon optimisation
From 2014.igem.org
(Difference between revisions)
| Line 29: | Line 29: | ||
<br> | <br> | ||
| - | <h1>Introduction: | + | <h1>Introduction: Codon Optimisation</h1> |
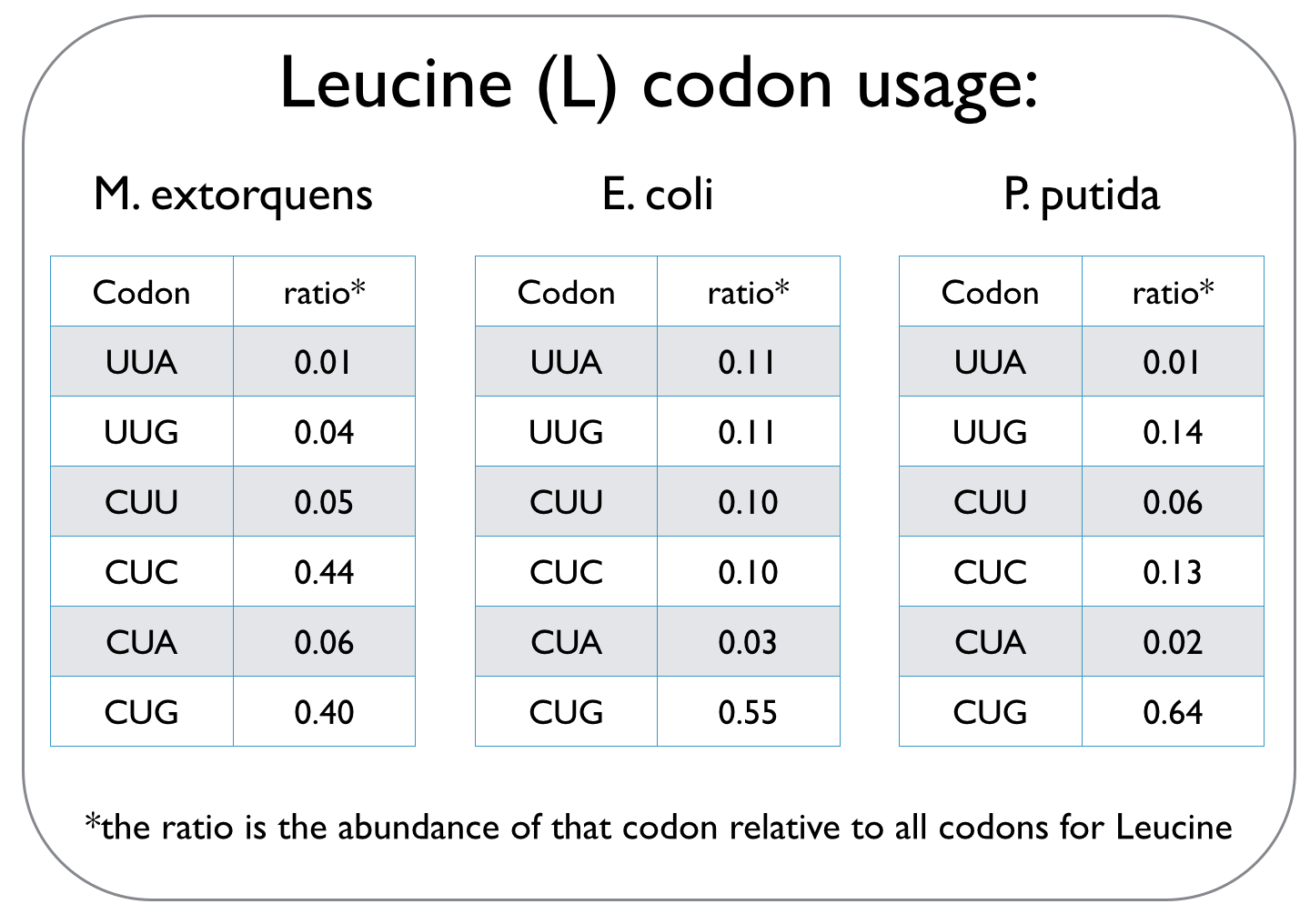
In our quest to optimise the process of DCM breakdown, the first step was to move the genes we were interested in into well-characterised host strains. The source bacterium Methylobacterium extorquens DM4 proved very difficult to grow in the lab, and took its leisurely time once it did decide to grow. It's also not a particularly well-studied bacterium, unlike E. coli and P. putida, which we decided to work with. This section explains the concept of codon optimisation, why we used it, and how we did so. <br><br> | In our quest to optimise the process of DCM breakdown, the first step was to move the genes we were interested in into well-characterised host strains. The source bacterium Methylobacterium extorquens DM4 proved very difficult to grow in the lab, and took its leisurely time once it did decide to grow. It's also not a particularly well-studied bacterium, unlike E. coli and P. putida, which we decided to work with. This section explains the concept of codon optimisation, why we used it, and how we did so. <br><br> | ||
</div> | </div> | ||
Revision as of 00:04, 18 October 2014
 "
"