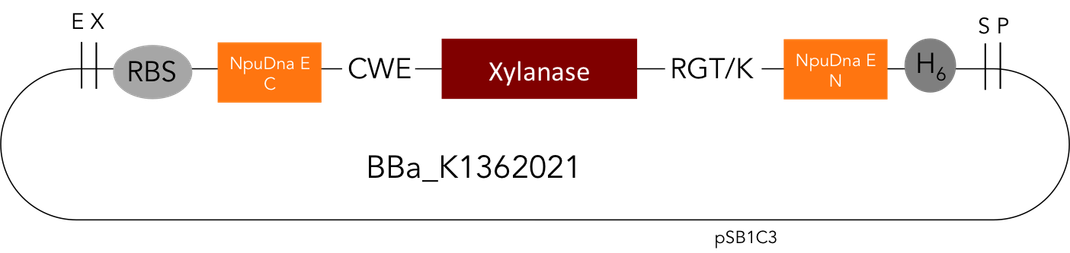
Team:Heidelberg/Parts/Part Improvement
From 2014.igem.org
(Created page with "{{:Team:Heidelberg/templates/wikipage_new| title=PART IMPROVEMENT | white=true | red-logo= | header-img= | body-style=background-color:black; | header-bg=#000000 | subtitle= This...") |
|||
| Line 12: | Line 12: | ||
header-bg=#000000 | header-bg=#000000 | ||
| | | | ||
| - | subtitle= | + | subtitle=Extending the Registry, One Step after Another |
| | | | ||
container-style=background-color:white; | container-style=background-color:white; | ||
Latest revision as of 08:50, 17 October 2014
– Extending the Registry, One Step after Another
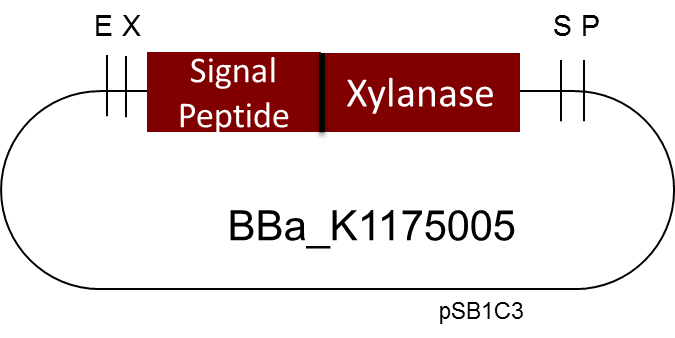
One of the central ideas of iGEM and Synthetic Biology is about modularity, which means standardization and reusing existing parts from the registry. During the course of our project we discovered a very interesting [http://parts.igem.org/Part:BBa_K1175005 Biobrick] by the Milwaukee iGEM Team 2013. It is a Xylanase from Bacillus subtilis; Xylanases degrade Xylan a major wood component. If you want to know more our work with Xylanases and their industrial applications please go to our page on Xylanases.
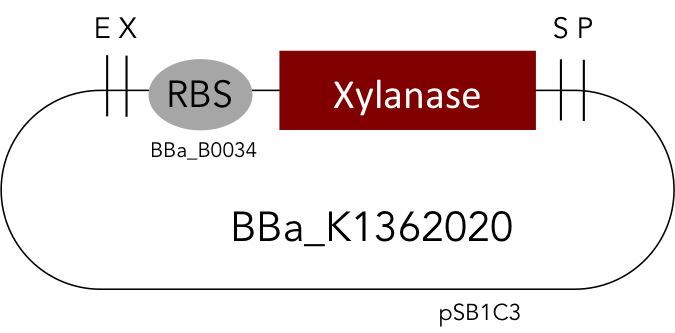
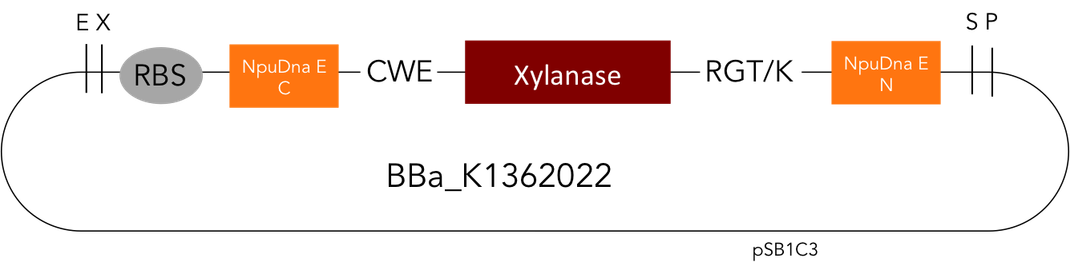
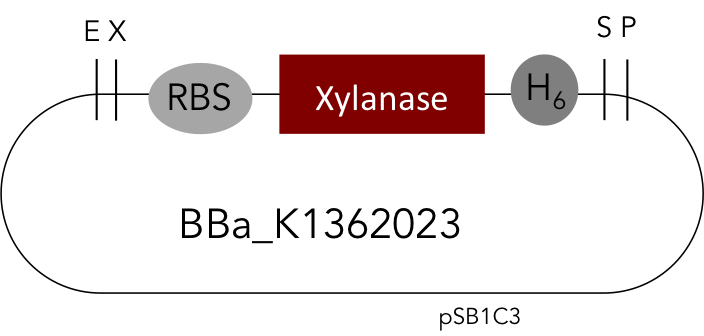
But as so often with existing things, the part by the Milwaukee Team wasn't a 100% suitable for our needs. That's why we decided to improve the part and its characterization. Although the original enzyme is produced by B. subtilis we were already working in our lab with E. coli. That's why decided to optimize the part for the expression in E. coli. The amino acids sequence of the Xylanase from B. subtilis has at the beginning a signal peptide, that is important for the extracellular export of the enzyme, afterwards the peptide is removed and only the mature peptide remains in the extracellular matrix. For the expression in E. coli we decided to remove this signaling part of the protein to make sure it stays intracellular, so the expression of circular and linear constructs could be compared. Furthermore we added a Ribosome Binding Site in front of the coding sequence as part of the BioBrick as it removes the need for an additional cloning step for an end user.
Plasmid Maps
[http://parts.igem.org/Part:BBa_K1175005 Linear Xylanase with Signal Peptide]
[http://parts.igem.org/Part:BBa_K1362020 Linear Xylanase]
[http://parts.igem.org/Part:BBa_K1362022 Circular Xylanase]
[http://parts.igem.org/Part:BBa_K1362023 Linear Xylanase with His-Tag]
[http://parts.igem.org/Part:BBa_K1362021 Circular Xylanase with His-Tag]
Additional Characterization
But we did not just add more parts to the registry, improving the characterization of existing parts is equally important. During the course of our project. As our project focused on heat stability, we determined the activity of our part in respect to its thermal stability.

Measurement of the fluorescence produced by the hydrolysis of commercially available substrate from the EnzChek® Ultra Xylanase Assay Kit by the linear, non-purified Xylanase (More information on assay).
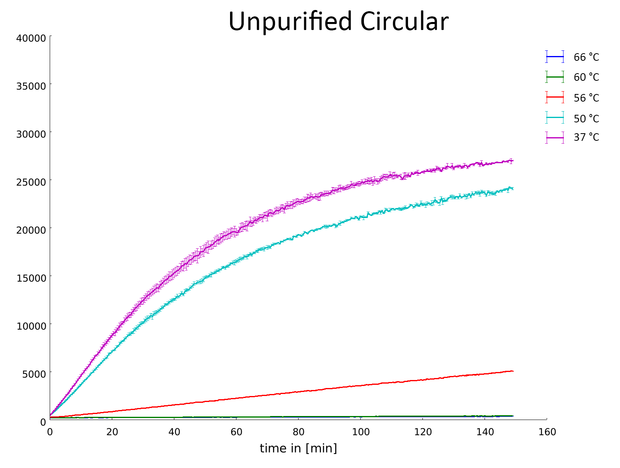
Measurement of the fluorescence produced by the hydrolysis of commercially available substrate from the EnzChek® Ultra Xylanase Assay Kit by the circular, non-purified Xylanase (More information on assay).
Our data indicates that the Xylanase has its optimal activity after a heatshock between 37 °C and 50 °C. At 60 °C and higher the Xylanase appears to be denatured and stopped working. This is an important characterization of the enzyme for its potential industrial applications.
 "
"