Team:Colombia/Interlab
From 2014.igem.org
Camilog137 (Talk | contribs) |
Camilog137 (Talk | contribs) |
||
| Line 146: | Line 146: | ||
<div class="accordion-inner"> | <div class="accordion-inner"> | ||
<h3>Measures</h3> | <h3>Measures</h3> | ||
| - | + | ||
The only difference between this part and the last one is the consitutive promoter, that in this case is J23115. The plasmid, RBS and stops are all the same. | The only difference between this part and the last one is the consitutive promoter, that in this case is J23115. The plasmid, RBS and stops are all the same. | ||
Revision as of 02:56, 6 August 2014
Interlab Study
This year Colombia Team-iGEM is participating in Interlab Study! We are glad of share our results and measures. You can find them in the tabs below, you can find the specifications of our equipments and the methodology we used as well.
Methods
Contents |
Strains and culture
All transformations were carried out in E. coli TOP10. This bacterium was cultured in LB (suplemented with the appropiate antibiotic for each case) at 37 °C.
Transformations and plasmidic DNA extration
This process was carried out as it is indicated in our protocols page.
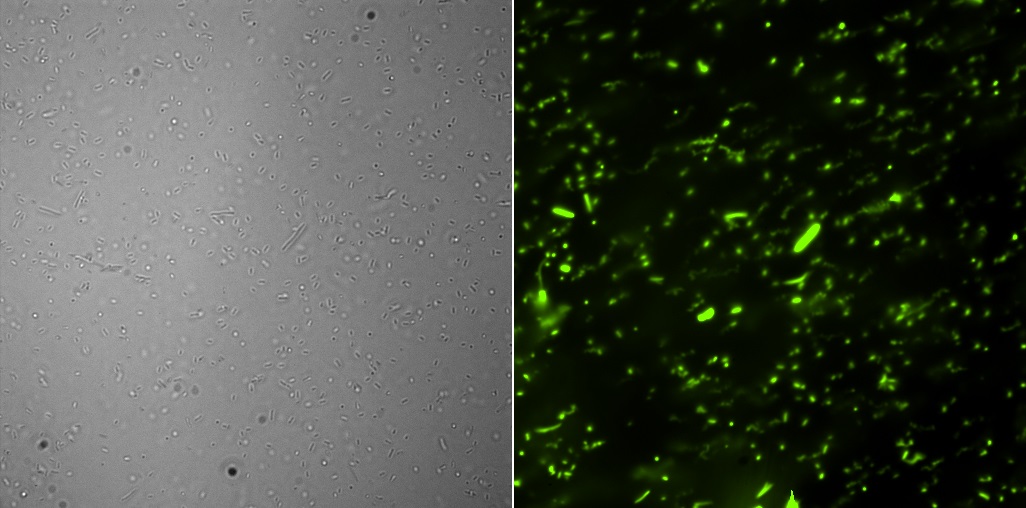
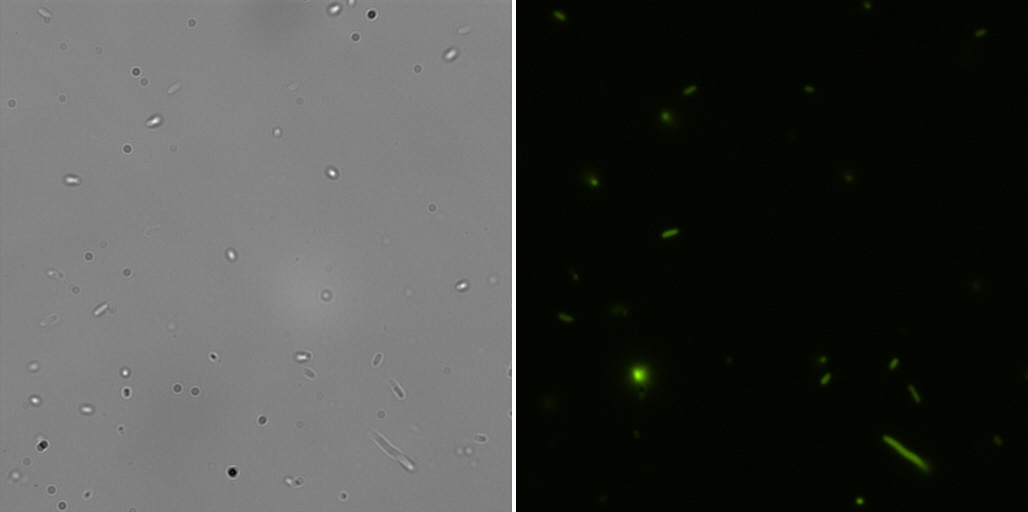
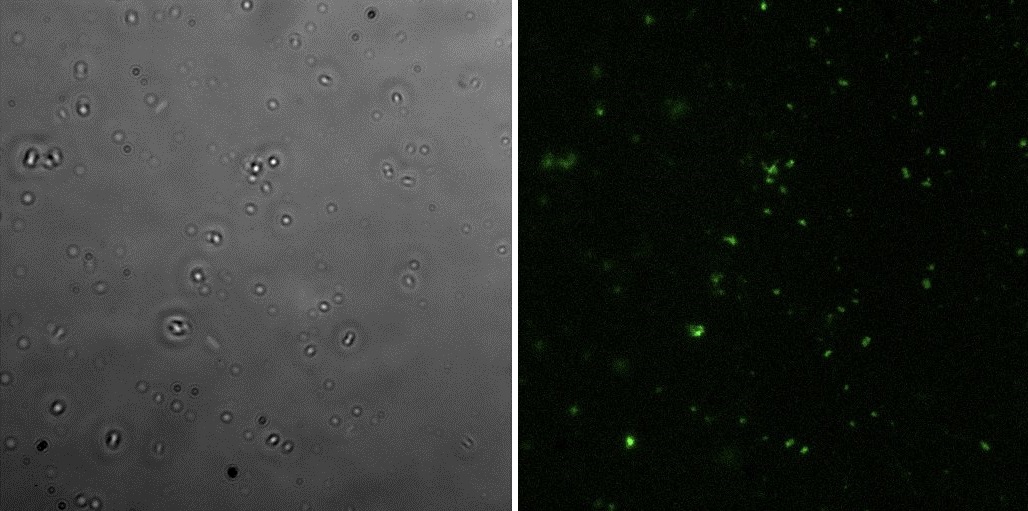
Fluorescence Microscopy
All the pictures shown here were taken using a Eclipse-Ti Nikon® ECLIPSE Ti inverted microscope equiped with an ANDOR iXon Ultra 897 camera. The filter used was FITC.
Fluorescence Microscopy
The fluorometric measures were taken using a DGEASGAES. The measures were carried out three times while 10 min with an ON culture diluted in a 1:10 proportion in 2mL of fresh LB, with a previous measuring of OD.
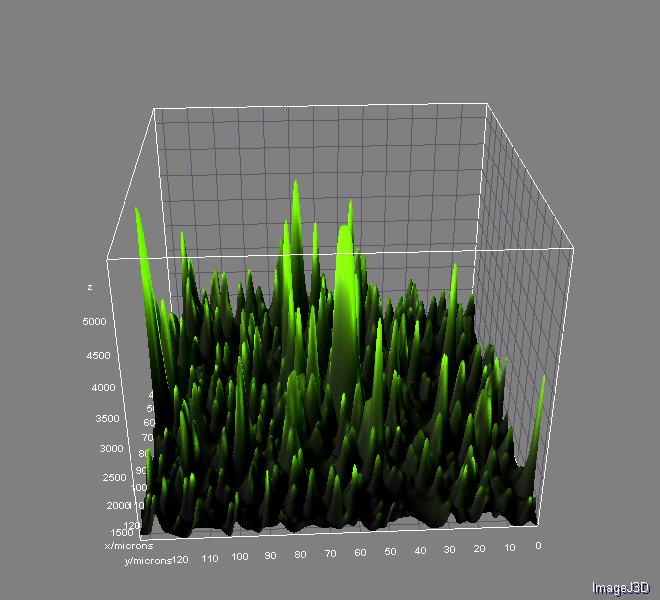
Image Analysis
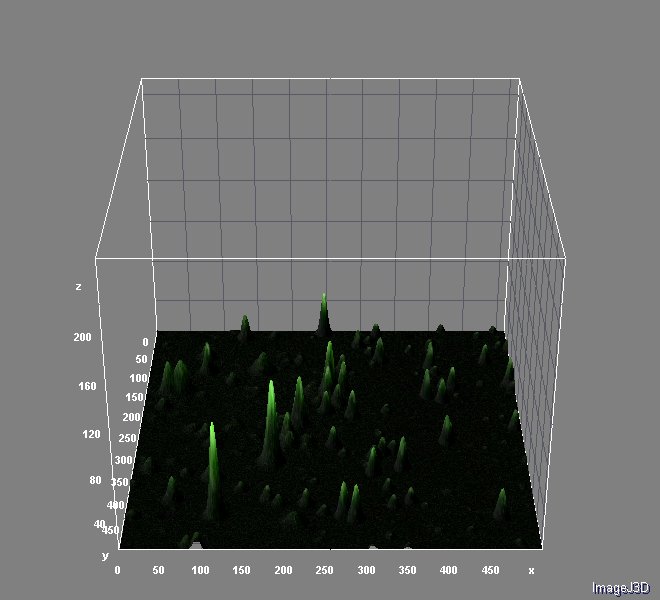
The image analysis were performed with ImageJ 1.46r using Nikon ND Reader and interactive 3D Surface Plot pluggins. We used Analyzing fluorescence microscopy images with ImageJ (Queen's University Belfast 2014) as guide. You can download this useful material here.
Validation
The following gel shows respectively the plasmids with the devices and the restriction profiles for each of the tree devices digested with EcoRI and PstI. For restrictions we obtained two bands (the plasmid and the device) with the expected sizes in all cases.
GELES :)
You can check the size of each device with the next table:
| Name | Backbone + device / bp | Device / bp | Linearized backbone / bp |
| BBa_I20260 | 3669 | 919 | 2750 |
| BBa_J23101 + BBa_E0240(B0032-E0040-B0015) | 2981 | 911 | 2070 |
| BBa_J23115 + BBa_E0240 (B0032-E0040-B0015) | 2981 | 911 | 2070 |
Measures
This part corresponds to GFP under the constitutive promoter J23101, with B0032 as RBS and two stops (B001 and B0010). The backbone of this already constructed device is pSB3K3 with kanamycin resistance.Fluorescence microscopy
Here are the images of the transformed cells with this device:
And the analysis for relative fluorescence among them performed from the last image above:
Fluorometry
dtgdxhedh
Measures
This part is identical to the last one, but built by us in pSB1C3.Fluorescence microscopy
Here are the images of the transformed cells with this device:
And the analysis for relative fluorescence among them performed from the last image above:
Fluorometry
Ejemplillo
 "
"