Team:Freiburg/Content/Results/Light system
From 2014.igem.org
Mirja Harms (Talk | contribs) |
Mirja Harms (Talk | contribs) |
||
| Line 67: | Line 67: | ||
<figure class="fig-full-width"> | <figure class="fig-full-width"> | ||
<a href="https://static.igem.org/mediawiki/2014/5/57/Freiburg2014-07-15_Rotlich_confocal_images.JPG"> <!-- ORGINAL --> | <a href="https://static.igem.org/mediawiki/2014/5/57/Freiburg2014-07-15_Rotlich_confocal_images.JPG"> <!-- ORGINAL --> | ||
| - | <img src="https://static.igem.org/mediawiki/2014/5/57/Freiburg2014-07-15_Rotlich_confocal_images.JPG | + | <img src="https://static.igem.org/mediawiki/2014/5/57/Freiburg2014-07-15_Rotlich_confocal_images.JPG"> <!-- Thumbnail --> |
</a> | </a> | ||
<figcaption> | <figcaption> | ||
| Line 106: | Line 106: | ||
</p> | </p> | ||
| - | <figure> | + | <figure class="fig-full-width"> |
<a href="https://static.igem.org/mediawiki/2014/3/31/Freiburg2014_Results_light_QR_96.jpg"> <!-- ORGINAL --> | <a href="https://static.igem.org/mediawiki/2014/3/31/Freiburg2014_Results_light_QR_96.jpg"> <!-- ORGINAL --> | ||
<img src="https://static.igem.org/mediawiki/2014/3/31/Freiburg2014_Results_light_QR_96.jpg"> <!-- Thumbnail --> | <img src="https://static.igem.org/mediawiki/2014/3/31/Freiburg2014_Results_light_QR_96.jpg"> <!-- Thumbnail --> | ||
Revision as of 17:29, 16 October 2014
The light system
One advantage of our system, the spatial resolution by infecting only distinct areas in a mammalian cell culture with our viral vector, was archived by the use of different light systems. We induced the expression of the mCAT-1 receptor in non-murine cell lines by illumination them with light of a special wave length. Since, the viral vector only infect cells that express the mCAT-1 receptor on their surfaces, we transferred genes into distinct areas of an otherwise homogenous cell layer using this principle.
Function of the Light Systems
As the function of the light system, we use, is really important for the function of our whole system, we tested two different light systems. The red light system is based on…. whereas the blue light system uses… blubb
Since, it is known that the red light system works better in chinese hamster ovary (CHO) cells, we used this cell line to test the function of this light system. In order to investigate the functionality of the blue light system, we tested both, CHO cells and human embryonic kidney (HEK293) cells. As reporter the gene seap was transfected together with the genes for the light systems (red light system: PKM006, PKM078; blue light system: PKM292, PKM297, PKM084). A SEAP-assay was performed after illumination. Regarding SEAP concentrations in the supernatant of each sample two important findings were archived. First, the blue light system works much more efficient than the red light system; and the blue light system works better with HEK cells than with CHO cells (Fig.1).Labjournal

Figure 2: Efficiency of the blue light system using different durations of illumination.
To test the efficiency of the light system light induced SEAP was used as a marker. A SEAP assay was performed after 24 hours of incubation. Cells were incubated with blue light for 1 hour, 2.5 hours and 5 hours.
The blue light system is very effective in activating an existing reporter, like the gene for seap, brought into the cell before illumination. As we know that the duration of illuminating the system with blue light (452 nm) is critical for its efficiency, we tested various time spans for illumination. Our results indicate that five hours of illumination lead to the highest activation level of SEAP expression.
Another important finding was the specificity of the blue light system. As we always have a dark control in our experiments, we determined almost no activation of the SEAP reporter in these samples leading to a very low background level in our system.
Light Induced Receptor
In order to generate viral induced patterns in a homogenous cell culture, we combined the spatial resolution of the light system with the specificity of viral receptor interactions. Since our viral vector is specific for the mCAT-1 receptor that is under normal conditions only expressed in murine cells, we linked this specific receptor to the blue light system or the red light system and transfected non-murine cells with this receptor combinations. Mammalian cells which contain the inducible receptor expressed mCAT-1 after illumination. Cells expressing the receptor are visible by co-expression of mCherry. They were infected with our viral vector leading to EGFP expression. Cells of the same cell culture that were not illuminated were not able to express the receptor leading to no infection with our viral vector.
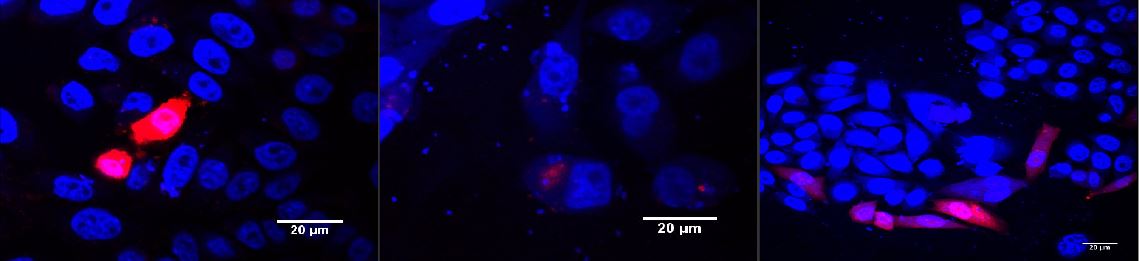
Red light induced receptor.
CHO cells were transfected with the red light system (PKM022) and the light induced receptor (p14rz_002). The receptor was labeled with mCherry; (left) after illumination with red light, (middle) incubation in the dark, (right) positive control.

Blue light induced receptor.
HEK cells were transfected with the blue light system (PKM292 and PKM297) and the light induced receptor (p14ls_003). The receptor was labeled with mCherry. Cells were infected with MuLV EGFP afterwards; (left) incubation in the dark, (middle) after illumination with blue light, (right) cells expressing the light induced receptor were infected with MuLV.
As we know that the transient mCAT-1 receptor that was not linked to a light system needs 24 hours for expression in HEK293 cells, we determined the expression time of the receptor that was induced by blue light after illumination for five hours. We used a receptor that was labeled with both, mCherry and an HA-tag (p14ls_003), for analysis with Western blot and fluorescence microscopy. The results show that the receptor had an expression peak at 24 hours after beginning of illumination.

Kinetik of the blue light induced receptor.
HEK cells were transfected with the blue light system (PKM292 and PKM297) and the light induced receptor (p14ls_003, mCherry labeled receptor). Pictures were taken after 12h, 15h, 18h and 24h.
SEAP as Reporter
In order to test the capability of the blue light system for pattern generation, we transfected HEK293 cells with the appropriate plasmids (PKM292 and PKM297) using seap as a reporter gene (PKM084). Cells were transfected in suspension and seeded on 96 well plates afterwards. Single wells of these plates were covered with a photo mask that prevents illumination with blue light. Since, all other wells on the plate were exposed to the blue light, SEAP expression was induced in these wells. SEAP that was secreted into the culture medium leads to colour changing from red to yellow after addition of pNPP (chromogenic substrate) by its phosphorylation.
First we generated a coarse pattern (fig.) realizing that by scattered light also covered wells close to illuminated wells were exposed to light. This was leading to an unclear pattern. However, we managed to give an idea of the desired pattern.
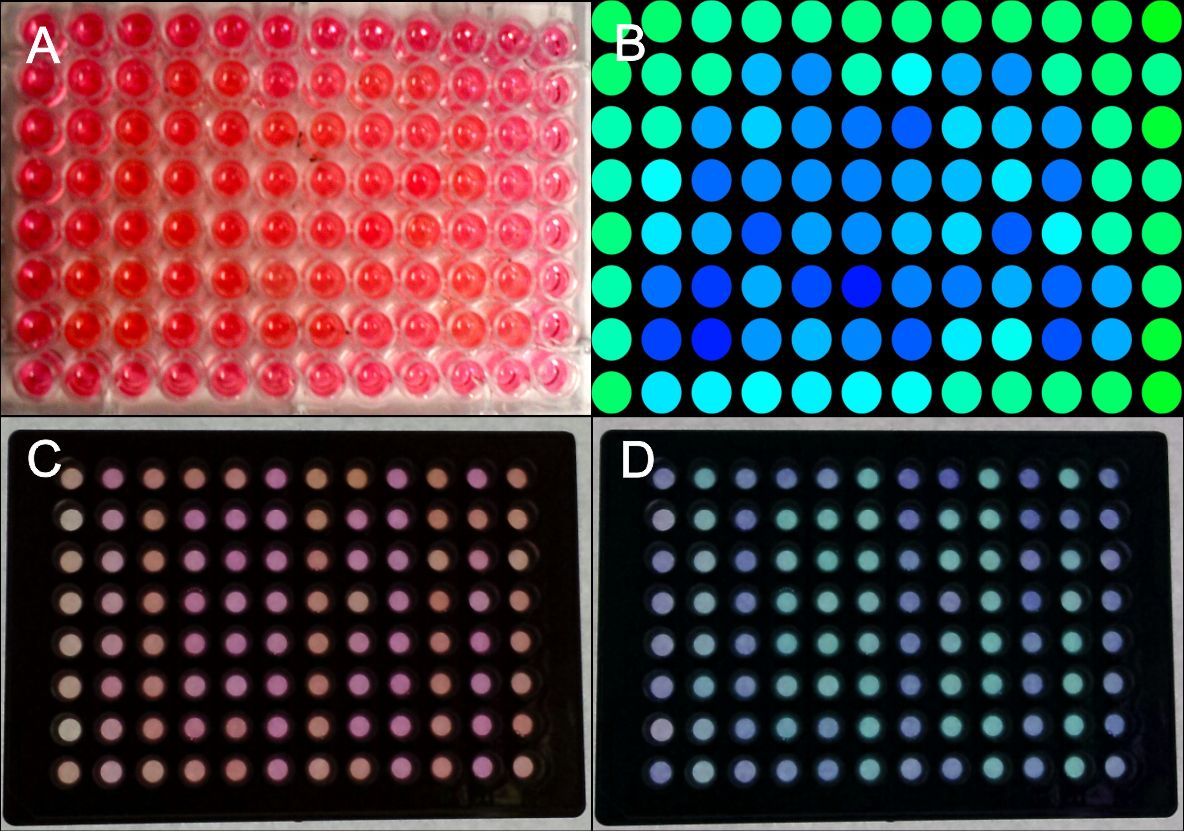
Generation of patterns on 96W plates.
HEK293 cells were transfected with the blue light system (PKM292, PKM297) and light induced SEAP as reporter (PKM084). Single wells were covered with a photo mask to prevent cell from light exposure. 24 hours after illumination with blue light a SEAP assay was performed leading to colour changing of wells containing SEAP. (A) Photo mask in heart form, (B) a pink colour was set to green and a yellow colour was set to blue at a computer, (C) the same experiment was performed with multiple well plates with separated wells, (D) the contrast an the colours of the wells were changed at a computer. All wells together are forming the word: IGEM.
Since, it was essential to completely prevent light exposure on covered plates we tested 96 well plates with optical separated wells. As shown in fig. we could generate patterns with sharp contours.
Colour changing of pNPP in cell culture medium could be measured at 405 nm in a plate reader. Calculating the gradient of light absorption we also generated patterns using a data matrix (Excel) by setting a distinct threshold, thus making pattern recognition easier.
 "
"
