Team:Aachen/Project/2D Biosensor
From 2014.igem.org
(→Principle of Operation) |
(→Principle of Operation) |
||
| Line 68: | Line 68: | ||
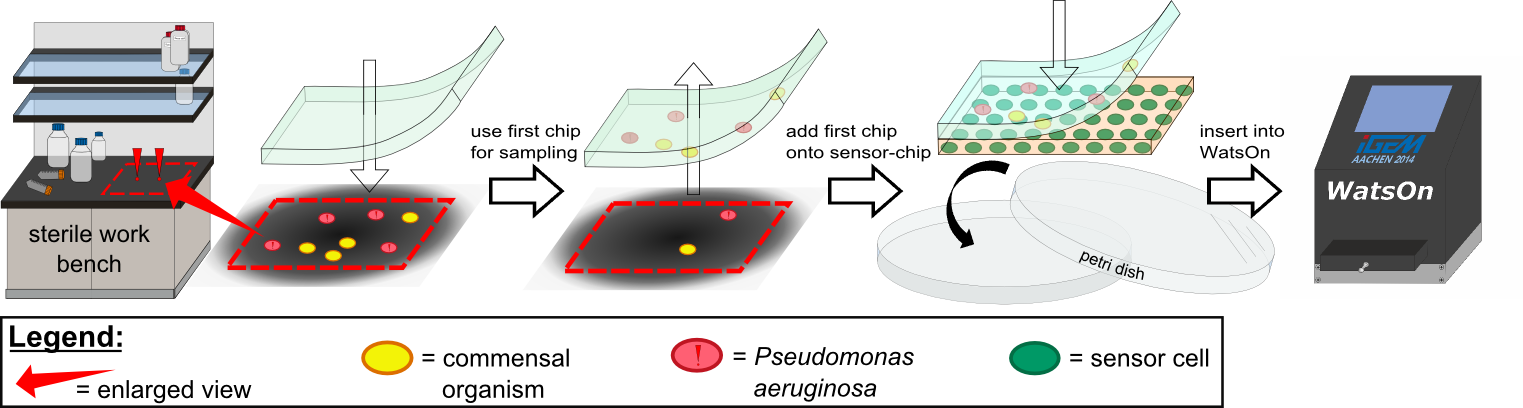
| - | The application of ''Cellock Holmes'' for detection of ''P. aeruginosa'' cells is | + | The application of ''Cellock Holmes'' for detection of ''P. aeruginosa'' cells is straightforward: Fist, a sampling chip is placed on a solid surface that is potentially contaminated with the pathogen. Second, the sampling chip is removed from the surface and put onto one of our sensor chips. Theorectically, the sensor chips could be directly used for sampling, however, this was avoided in our project to '''match [https://2014.igem.org/Team:Aachen/Safety biosafety regulations]''' and to prevent the spread of GMOs into the environment. Third, the two layered chip-stack is put into a petri dish which is inserted into our measurement device [https://2014.igem.org/Team:Aachen/Project/Measurement_Device ''WatsOn''] for evalutation. |
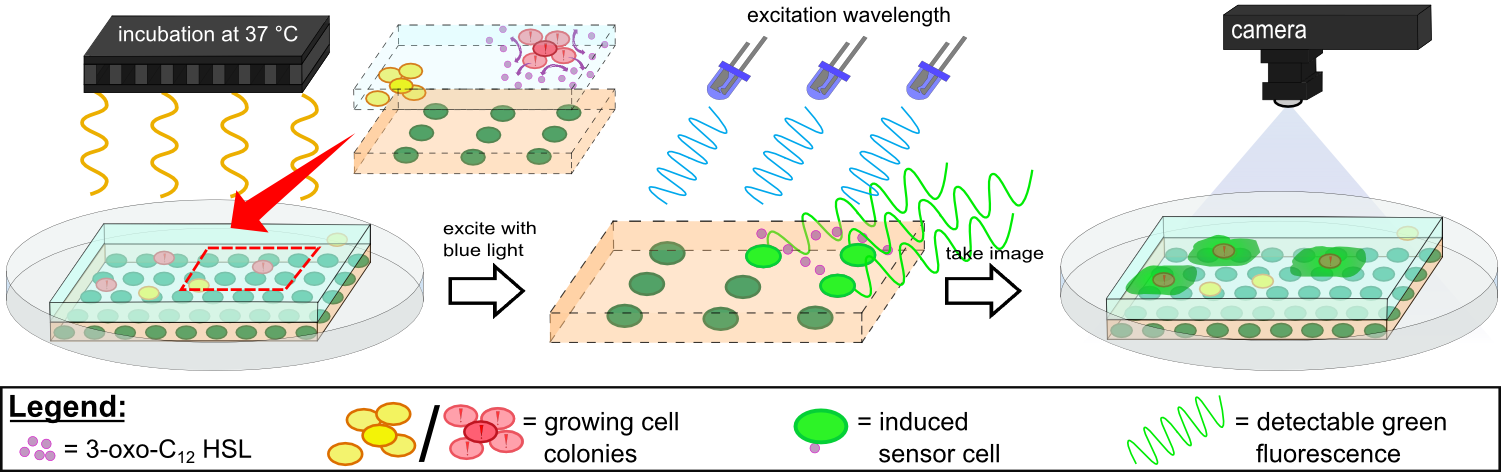
{{Team:Aachen/Figure|Aachen 14-10-14 Flowsheet OD-device part2 ipo.png|title=Mode of action inside WatsOn.|width=1000px}} | {{Team:Aachen/Figure|Aachen 14-10-14 Flowsheet OD-device part2 ipo.png|title=Mode of action inside WatsOn.|width=1000px}} | ||
| - | Inside ''WatsOn'' the chips are incubated at 37 °C and populations of microorganisms stuck on the sampling chip start to multiply. ''P. aeruginosa'' secrets an increasing number of quorum sensing molecules in growing cell colonies | + | Inside ''WatsOn'', the chips are incubated at 37 °C and populations of microorganisms stuck on the sampling chip start to multiply. ''P. aeruginosa'' secrets an increasing number of quorum sensing molecules in growing cell colonies which are taken up by our sensor cells where they induce a fluorescence signal. In case of ''P. aeruginosa'', we focused on quorum sensing molecules called N-3-oxo-dodecanoyl-L-homoserine lactones, or 3-oxo-C-12-HSLs for short, while our sensor cells contain of genetically modified ''E. coli'' cells which are able to elecit a '''fluorescent response to autoinducers'''. The 3-oxo-C-12-HSL detection system incorporated into the sensor cells is explained in more detail in the [https://2014.igem.org/Team:Aachen/Project/FRET_Reporter REACh Construct] section. |
| - | Inside ''WatsOn' the chips can be illuminated with blue light at any time, whereby a picture of the chip is taken. The software ''Measurarty'' then analyzes any fluorescent signal. <!-- Depending on the intensity of the signal and the size of the spot, ''Cellock Holmes'' can '''calculate concentration and distribution of ''P. aeruginosa'' ''' on the sampled surface. --> | + | Inside ''WatsOn' the chips can be illuminated with blue light at any time, whereby a picture of the chip is taken. The software ''Measurarty'' then analyzes any fluorescent signal. <!-- Depending on the intensity of the signal and the size of the spot, ''Cellock Holmes'' can '''calculate concentration and distribution of ''P. aeruginosa'' ''' on the sampled surface. --> |
</center> | </center> | ||
| Line 85: | Line 85: | ||
<span class="anchor" id="biosensormolecularapproach"></span> | <span class="anchor" id="biosensormolecularapproach"></span> | ||
| - | For our biosensor, our team genetically modified ''E. coli'' cells to be able to elecit a '''fluorescent response to autoinducers''' produced by the pathogen ''Pseudomonas aeruginosa'' during quorum sensing. In the case of ''P. aeruginosa'', these autoinducers are N-3-oxo-dodecanoyl-L-homoserine lactone, or 3-oxo-C-12-HSL for short. The quorum sensing system of this pathogen contains the '''LasR activator''' which binds 3-oxo-C-12-HSL, and the '''LasI promoter''', which is activated by the LasR-HSL complex. Both LasR activator and LasI promoter are available as BioBricks [http://parts.igem.org/Part:BBa_C0179 C0179] and [http://parts.igem.org/Part:BBa_J64010 J64010]. | + | For our biosensor, our team genetically modified ''E. coli'' cells to be able to elecit a '''fluorescent response to autoinducers''' produced by the pathogen ''Pseudomonas aeruginosa'' during quorum sensing. In the case of ''P. aeruginosa'', these autoinducers are N-3-oxo-dodecanoyl-L-homoserine lactone, or 3-oxo-C-12-HSL for short. The quorum sensing system of this pathogen contains the '''LasR activator''' which binds 3-oxo-C-12-HSL, and the '''LasI promoter''', which is activated by the LasR-HSL complex. Both LasR activator and LasI promoter are available as BioBricks [http://parts.igem.org/Part:BBa_C0179 C0179] and [http://parts.igem.org/Part:BBa_J64010 J64010]. |
As a reporter gene, we use '''GFP'''. However, expression of GFP is not simply controlled through the LasI promoter activity in our approach. Instead, our sensor cells contain genes for a constitutively expressed fusion protein consisting of GFP and a dark quencher, and an '''HSL-inducible protease'''. We use the REACh protein as dark quencher for GFP and the TEV protease to cleave the complex; [https://2014.igem.org/Team:Aachen/Project/FRET_Reporter here] you can read more about the REACh construct and the TEV protease. | As a reporter gene, we use '''GFP'''. However, expression of GFP is not simply controlled through the LasI promoter activity in our approach. Instead, our sensor cells contain genes for a constitutively expressed fusion protein consisting of GFP and a dark quencher, and an '''HSL-inducible protease'''. We use the REACh protein as dark quencher for GFP and the TEV protease to cleave the complex; [https://2014.igem.org/Team:Aachen/Project/FRET_Reporter here] you can read more about the REACh construct and the TEV protease. | ||
| Line 91: | Line 91: | ||
{{Team:Aachen/Figure|align=center|Aachen_REACh_approach.png|title=Our novel biosensor approach|subtitle=Expression of the TEV protease is induced by HSL. The protease cleaves the GFP-REACh fusion protein to elecit a fluorescence response.|width=500px}} | {{Team:Aachen/Figure|align=center|Aachen_REACh_approach.png|title=Our novel biosensor approach|subtitle=Expression of the TEV protease is induced by HSL. The protease cleaves the GFP-REACh fusion protein to elecit a fluorescence response.|width=500px}} | ||
| - | When ''P. aeruginosa cells'' are stuck on our agar chip and come close to our sensor cells, the latter take up the HSL molecules secreted by the pathogens. Inside the sensor cells, the autoinducer binds to the LasR gene product and activate the expression of the TEV protease. The protease then cleaves the GFP-REACh construct. When '''illuminated with light of 480 nm''', the excitation wavelenght of GFP, our sensor cells in the vicinity of ''P. aeruginosa'' give a '''fluorescence signal'''. On the other hand, sensor cells that were not anywhere close to the pathogens do not express the protease. Therefore, the GFP will still be attached to the dark quencher in these cells, and no fluorescence is produced. | + | When ''P. aeruginosa cells'' are stuck on our agar chip and come close to our sensor cells, the latter take up the HSL molecules secreted by the pathogens. Inside the sensor cells, the autoinducer binds to the LasR gene product and activate the expression of the TEV protease. The protease then cleaves the GFP-REACh construct. When '''illuminated with light of 480 nm''', the excitation wavelenght of GFP, our sensor cells in the vicinity of ''P. aeruginosa'' give a '''fluorescence signal'''. On the other hand, sensor cells that were not anywhere close to the pathogens do not express the protease. Therefore, the GFP will still be attached to the dark quencher in these cells, and no fluorescence is produced. |
Revision as of 07:53, 16 October 2014
|
|
|
 "
"