Team:Aachen/Notebook/Wetlab/October
From 2014.igem.org
(Difference between revisions)
Aschechtel (Talk | contribs) (→10th) |
(→3rd) |
||
| (55 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| + | {{CSS/Main}} | ||
| + | {{Team:Aachen/Stylesheet}} | ||
{{Team:Aachen/Header}} | {{Team:Aachen/Header}} | ||
| + | <html> | ||
<center> | <center> | ||
| - | + | <ul class="menusmall-grid"> | |
| - | + | ||
| - | + | <!-- <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | |
| + | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/March" style="color:black"> | ||
| + | <div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to March</div></div> | ||
| + | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/7/7a/Aachen_14-10-10_March_iFG.png); norepeat scroll 0% 0% transparent; background-size:100%; height:100px; width: 100px;"> | ||
| + | </div> | ||
| + | </a> | ||
| + | </li> --> | ||
| + | |||
| + | <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | ||
| + | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/April" style="color:black"> | ||
| + | <div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to April</div></div> | ||
| + | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/2/2d/Aachen_14-10-10_April_iFG.png); norepeat scroll 0% 0% transparent; background-size:100%; height:100px; width: 100px;"> | ||
| + | </div> | ||
| + | </a> | ||
| + | </li> | ||
| + | |||
| + | <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | ||
| + | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/May" style="color:black"> | ||
| + | <div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to May</div></div> | ||
| + | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/6/67/Aachen_14-10-10_May_iFG.png); norepeat scroll 0% 0% transparent; background-size:100%; height:100px; width: 100px;"> | ||
| + | </div> | ||
| + | </a> | ||
| + | </li> | ||
| + | |||
| + | <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | ||
| + | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/June" style="color:black"> | ||
| + | <div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to June</div></div> | ||
| + | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/1/1d/Aachen_14-10-10_June_iFG.png); norepeat scroll 0% 0% transparent; background-size:100%; height:100px; width: 100px;"> | ||
| + | </div> | ||
| + | </a> | ||
| + | </li> | ||
| + | |||
| + | <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | ||
| + | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/July" style="color:black"> | ||
| + | <div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to July</div></div> | ||
| + | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/1/19/Aachen_14-10-10_July_iFG.png); norepeat scroll 0% 0% transparent; background-size:100%; height:100px; width: 100px;"> | ||
| + | </div> | ||
| + | </a> | ||
| + | </li> | ||
| + | |||
| + | <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | ||
| + | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/August" style="color:black"> | ||
| + | <div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to August</div></div> | ||
| + | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/4/4e/Aachen_14-10-10_August_iFG.png); norepeat scroll 0% 0% transparent; background-size:100%; height:100px; width: 100px;"> | ||
| + | </div> | ||
| + | </a> | ||
| + | </li> | ||
| + | |||
| + | <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | ||
| + | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/September" style="color:black"> | ||
| + | <div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to September</div></div> | ||
| + | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/d/d4/Aachen_14-10-10_September_iFG.png); norepeat scroll 0% 0% transparent; background-size:100%; height:100px; width: 100px;"> | ||
| + | </div> | ||
| + | </a> | ||
| + | </li> | ||
| + | |||
| + | <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | ||
| + | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/October" style="color:black"> | ||
| + | <div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to October</div></div> | ||
| + | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/6/60/Aachen_14-10-10_October_iFG.png); norepeat scroll 0% 0% transparent; background-size:100%; height:100px; width: 100px;"> | ||
| + | </div> | ||
| + | </a> | ||
| + | </li> | ||
| + | |||
| + | </ul> | ||
</center> | </center> | ||
| + | </html> | ||
= October = | = October = | ||
== 1st == | == 1st == | ||
| - | * Prepartations for sensor-chip production the following day (2014-10-02) was done accoringly to the [https://2014.igem.org/Team:Aachen/Notebook/Protocols | + | * Prepartations for sensor-chip production the following day (2014-10-02) was done accoringly to the [https://2014.igem.org/Team:Aachen/Notebook/Protocols/detection sensor-chip manufacturing protocol]: |
| - | ** At 18:30 we prepared over-night cultures from K1319042, B0015 and K131026 by inoculating 250 ml Erlenmeyer flasks each containing 50 ml LB medium . The flasks were incubated for ~12 hours at | + | ** At 18:30 we prepared over-night cultures from K1319042, B0015 and K131026 by inoculating 250 ml Erlenmeyer flasks each containing 50 ml LB medium . The flasks were incubated for ~12 hours at 37°C on a shaker. |
== 2nd == | == 2nd == | ||
| Line 18: | Line 86: | ||
* made precultures and a master plate of 6 colonies of K3139008 in psB1C3 in NEB10β cells that had been plated at 5:30 this morning. | * made precultures and a master plate of 6 colonies of K3139008 in psB1C3 in NEB10β cells that had been plated at 5:30 this morning. | ||
| - | * Production of sensor-chips was done accordingly to [https://2014.igem.org/Team:Aachen/Notebook/Protocols | + | * Production of sensor-chips was done accordingly to [https://2014.igem.org/Team:Aachen/Notebook/Protocols/detection sensor-chip manufacturing protocol]. Briefly: |
| - | ** At 10:00 we prepared 150 ml 1.5 | + | ** At 10:00 we prepared 150 ml 1.5% (w/v) LB+agarose solution. The LB+agarose solution was autoclaved and subsequently tempered to 45°C. Precultures (50 ml each) of K1319042, B0015 and K131026 were spun down at 3000 g for 10 min at 21°C and re-suspended in 1 ml pretempered (21°C) LB medium. The re-suspended cultures were mixed with 50 ml LB+agarose and poured onto three sensor-chip-templates (one template per culture). Sensor chips were cut out from the template and incubated at 37°C for 1 h. |
** K1319042 and B0015 were induced with 0.2 µl IPTG (100 mM) subsequently to incubation and K131026 was induced with 0.2 µl homoserinlacton stock solution (500 µg/ml) 30 minutes after induction of the K1319042 and B0015. The induced sensor-chips were read out every 30 minutes for 180 minutes in total. An additional readout was conducted 285 minutes post induction. The readout was done at 450 nm and 480 nm wavelength. | ** K1319042 and B0015 were induced with 0.2 µl IPTG (100 mM) subsequently to incubation and K131026 was induced with 0.2 µl homoserinlacton stock solution (500 µg/ml) 30 minutes after induction of the K1319042 and B0015. The induced sensor-chips were read out every 30 minutes for 180 minutes in total. An additional readout was conducted 285 minutes post induction. The readout was done at 450 nm and 480 nm wavelength. | ||
<center> | <center> | ||
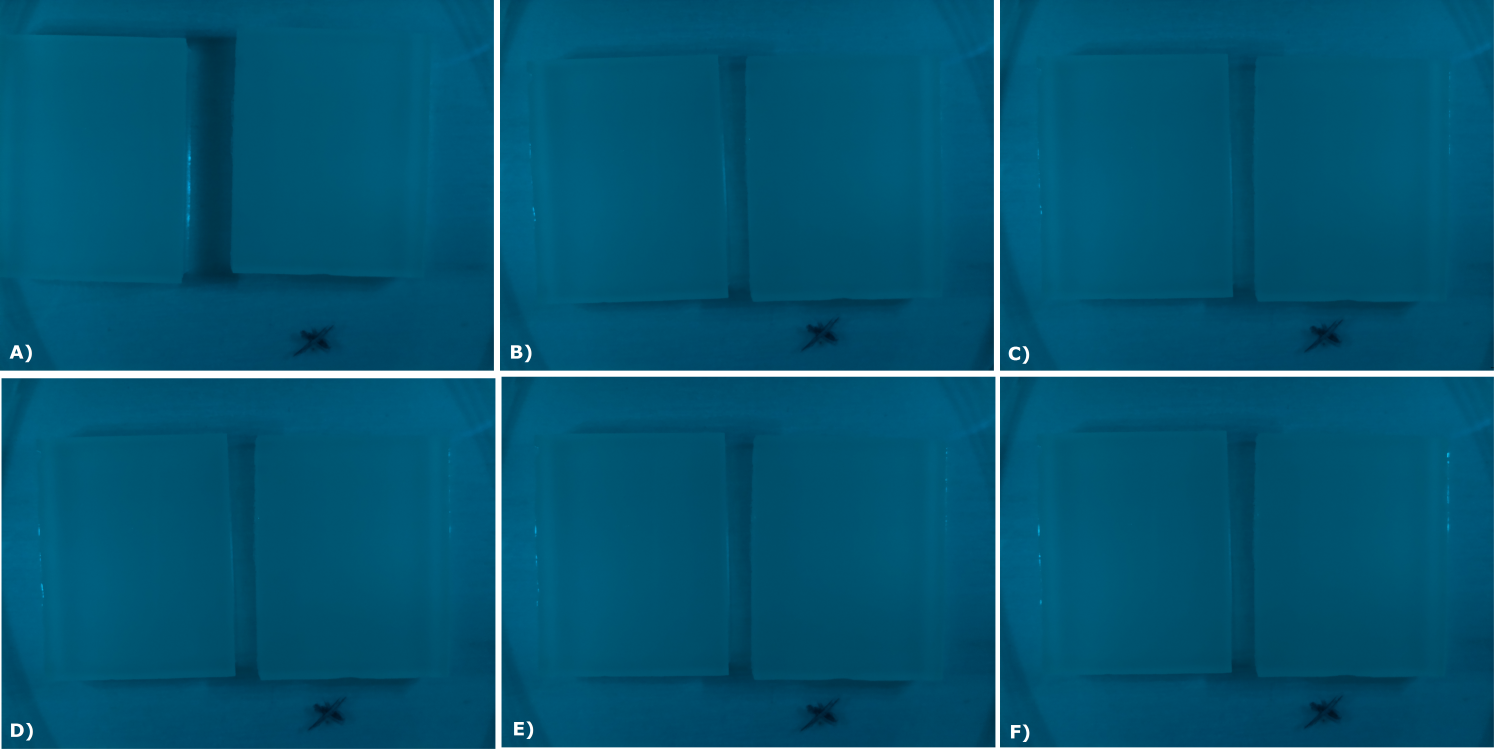
| - | {{Team:Aachen/Figure|Aachen_02_10_2014_B0015_serie.png|title=Sensor Chips with B0015 in NEB in LB (480 nm): first try | + | {{Team:Aachen/Figure|Aachen_02_10_2014_B0015_serie.png|title=Sensor Chips with B0015 in NEB in LB (480 nm): first try with final device|subtitle=Sensor chips with B0015 in NEB in LB medium with 1,5% agarose, right chip induced. A) befor induction with 0.2 µl of 100 mM IPTG B) 0.5 h after induction C) 1 h after induction D) 1.5 h after induction E) 2 h after induction F) 2.5 h after induction|width=900px}} |
</center> | </center> | ||
| Line 72: | Line 140: | ||
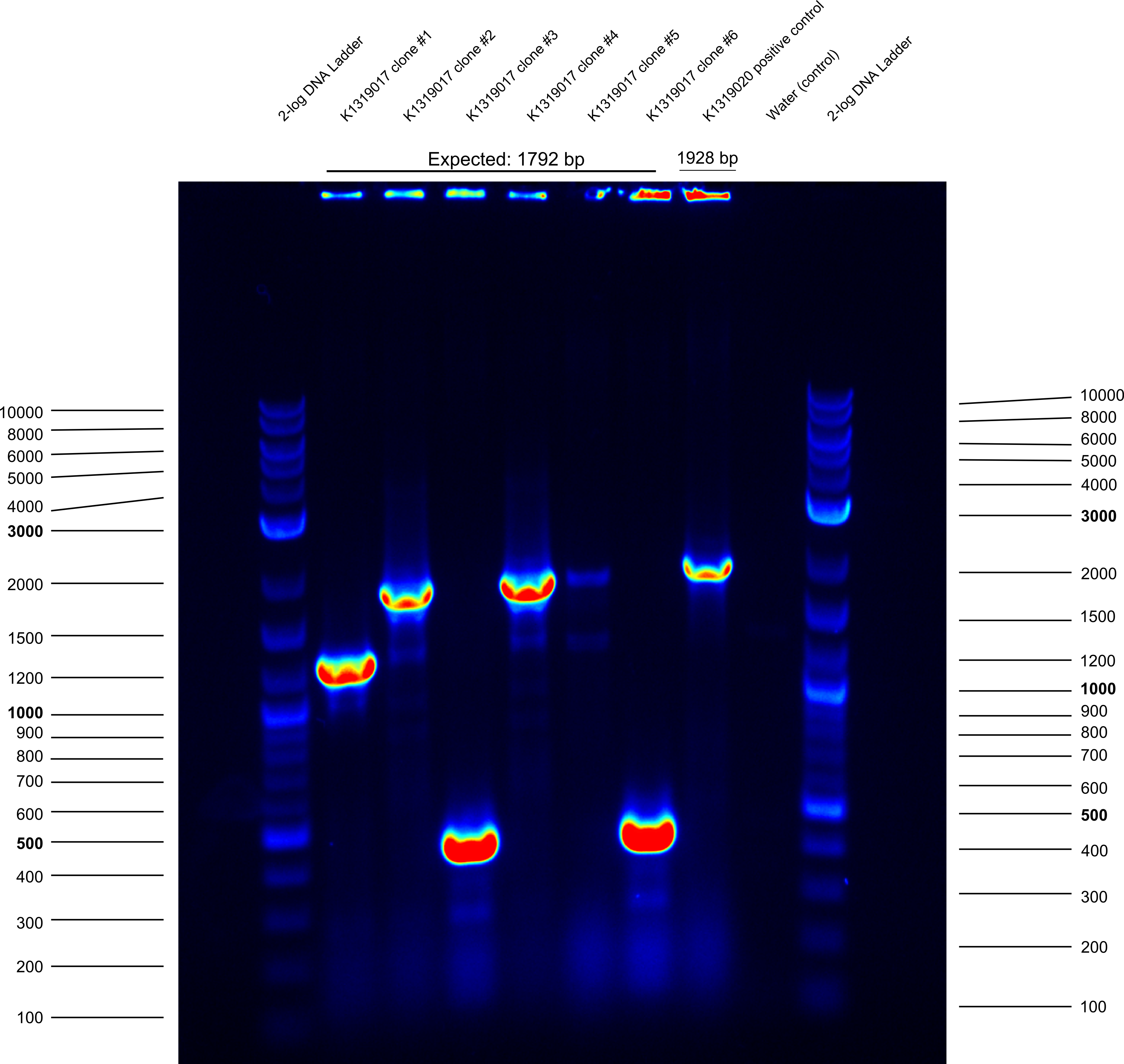
* did plasmid prep and cryo of clones #2 and #4 of K1319017 | * did plasmid prep and cryo of clones #2 and #4 of K1319017 | ||
| - | * new plasmid backbone of pSB1C3 was made using the [http://parts.igem.org/Help:Protocols/Linearized_Plasmid_Backbones | + | * new plasmid backbone of pSB1C3 was made using the [http://parts.igem.org/Help:Protocols/Linearized_Plasmid_Backbones Standard Protocol]. |
* transformation of K1319008 clone #1 (from BL21) and K1319013 into BL21 (two plasmids in one cell). | * transformation of K1319008 clone #1 (from BL21) and K1319013 into BL21 (two plasmids in one cell). | ||
| Line 78: | Line 146: | ||
* transformation of K1319008 clone #1 (from BL21) and k1319014 into BL21 (two plasmids in one cell). | * transformation of K1319008 clone #1 (from BL21) and k1319014 into BL21 (two plasmids in one cell). | ||
| - | * OD measurements of three biological triplicates from ''E. coli'' BW21 113, ''P. putida'' and ''S. cerevisiae''. Measurement as an analytic triplicate in the spectrophotometer (absorbance and transmission) and our own OD/F | + | * OD measurements of three biological triplicates from ''E. coli'' BW21 113, ''P. putida'' and ''S. cerevisiae''. Measurement as an analytic triplicate in the spectrophotometer (absorbance and transmission) and our own OD/F Device. |
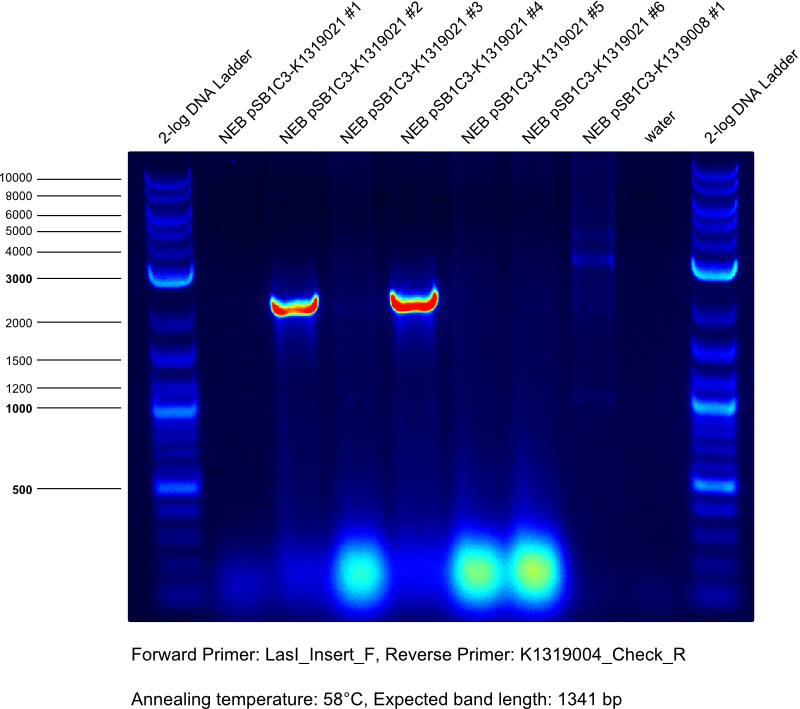
* Gibson assembly of K1319021 | * Gibson assembly of K1319021 | ||
| Line 84: | Line 152: | ||
** template Insert: LasI gene synthesis | ** template Insert: LasI gene synthesis | ||
| - | * At 19:00 we measured OD (our OD-device), absorption (spectrophotometer) and transmission (spectrophotometer) for 19 diltuions in the range of 2.5-100 | + | * At 19:00 we measured OD (our OD-device), absorption (spectrophotometer) and transmission (spectrophotometer) for 19 diltuions in the range of 2.5-100% from yeast (''Saccharomyces cerevisiae'') and ''P. putida'' liquid cultures. Measurements were conducted in biological as well as technical triplicates. Aim of this experiment was the comparison of our OD-device to commonly used devices in terms of OD determination. |
| - | * Prepartations for sensor-chip production the following day (2014-10-04) were done accordingly to the [https://2014.igem.org/Team:Aachen/Notebook/Protocols | + | * Prepartations for sensor-chip production the following day (2014-10-04) were done accordingly to the [https://2014.igem.org/Team:Aachen/Notebook/Protocols/detection sensor-chip manufacturing protocol]: |
| - | ** At 22:00 we prepared over-night cultures from B0015, K1319017 and K131026 by inoculating 250 mL Erlenmeyer flasks each containing 50 mL LB medium for sensor-chip manufacturing the next day. The flasks were incubated for ~12 hours at | + | ** At 22:00 we prepared over-night cultures from B0015, K1319017 and K131026 by inoculating 250 mL Erlenmeyer flasks each containing 50 mL LB medium for sensor-chip manufacturing the next day. The flasks were incubated for ~12 hours at 37°C on a shaker. |
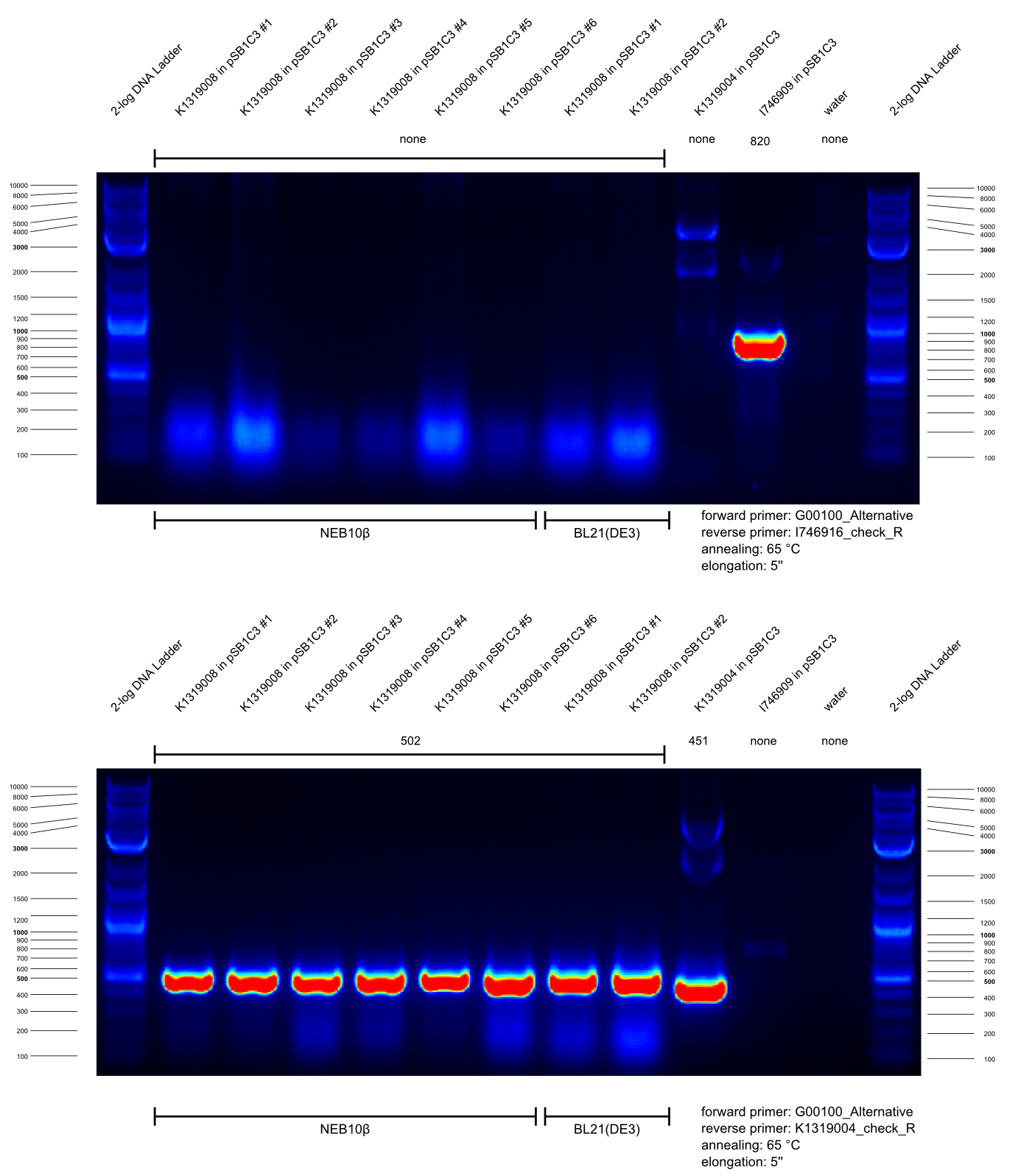
* Our BioBrick K1319021 enables expression of the TEV protease inducible by the autoinducers of ''Pseumomonas aeruginosa''. In order to construct this BioBrick, a Gibson assembly of K1319008 (IPTG-inducible expression of TEV protease) and the synthezised composite HSL-promotor J23101.B0032.C0079.B0015.J64010.B0034 activated by the respective autoinducers (HomoSerineLactones) was perfomed. Prior to this, we used two PCRs to linearize pSB1C3-K1319008, serving as backbone for Gibson assembly, and cutting out J23101.B0032.C0079.B0015.J64010.B0034 as well as adding adequate overlapping sequences. | * Our BioBrick K1319021 enables expression of the TEV protease inducible by the autoinducers of ''Pseumomonas aeruginosa''. In order to construct this BioBrick, a Gibson assembly of K1319008 (IPTG-inducible expression of TEV protease) and the synthezised composite HSL-promotor J23101.B0032.C0079.B0015.J64010.B0034 activated by the respective autoinducers (HomoSerineLactones) was perfomed. Prior to this, we used two PCRs to linearize pSB1C3-K1319008, serving as backbone for Gibson assembly, and cutting out J23101.B0032.C0079.B0015.J64010.B0034 as well as adding adequate overlapping sequences. | ||
| Line 99: | Line 167: | ||
| Initial denaturation || 05:00 || 98 || 05:00 || 98 || | | Initial denaturation || 05:00 || 98 || 05:00 || 98 || | ||
|- | |- | ||
| - | | Denaturation || 00:30 || 98 || 00:30 || 98 || rowspan="3" | '''30 cycles''' | + | | '''Denaturation''' || '''00:30''' || '''98''' || '''00:30''' || '''98''' || rowspan="3" | '''30 cycles''' |
|- | |- | ||
| - | | Annealing || 00:30 || 55 || 00:30 || 51 | + | | '''Annealing''' || '''00:30''' || '''55''' || '''00:30''' || '''51''' |
|- | |- | ||
| - | | Elongation || 01:35 || 72 || 00:37 || 72 | + | | '''Elongation''' || '''01:35''' || '''72''' || '''00:37''' || '''72''' |
|- | |- | ||
| Final elongation || 05:00 || 72 || 05:00 || 72 || | | Final elongation || 05:00 || 72 || 05:00 || 72 || | ||
| Line 139: | Line 207: | ||
* shake flask experiments with K1319008 (clone #1), K1319013 + K1319008 (clone #2) and K1319014 + K1319008 (clone #2) in LB (2 flasks each); inoculation with 50 µL preculture and inducing with iPTG at OD of 1.5. | * shake flask experiments with K1319008 (clone #1), K1319013 + K1319008 (clone #2) and K1319014 + K1319008 (clone #2) in LB (2 flasks each); inoculation with 50 µL preculture and inducing with iPTG at OD of 1.5. | ||
| - | * Production of sensor-chips was done accordingly to the [https://2014.igem.org/Team:Aachen/Notebook/Protocols | + | * Production of sensor-chips was done accordingly to the [https://2014.igem.org/Team:Aachen/Notebook/Protocols/detection sensor-chip manufacturing protocol]: |
** At 13:30 we prepared sensor chips from pre-cultures of B0015, K1319017 and K131026. | ** At 13:30 we prepared sensor chips from pre-cultures of B0015, K1319017 and K131026. | ||
| - | ** Subsequently to 1 h icubation at | + | ** Subsequently to 1 h icubation at 37°C B0015, K1319017 and K131026 were induced with 0.2 µL homoserinlacton stock solution (500 µg/ml). The induced sensor-chips were read out every 30 minutes for 240 minutes in total. Readout was conducted at 450 nm and 480 nm wavelength. An additional readout was conducted after 12 hours. |
* At 17:00 we prepared 4 liquid cultures from the K1319010_pSB3K3 master plate (clone#1) in 5 mL LB-medium, each. The liquid cultures were prepared in order to create cryo stocks from K1319010-pSB3K3. Kanamycin was added to the liquid cultures as antibiotic at an concentration of 1 µL/mL. | * At 17:00 we prepared 4 liquid cultures from the K1319010_pSB3K3 master plate (clone#1) in 5 mL LB-medium, each. The liquid cultures were prepared in order to create cryo stocks from K1319010-pSB3K3. Kanamycin was added to the liquid cultures as antibiotic at an concentration of 1 µL/mL. | ||
| - | * At 18:00 we prepared a master plate (LB+C) and corresonding liquid cultures from 6 clones of ''E.coli'' NEB10B k1319021-psB1C3. Liquid cultures and master plate were incubated at | + | * At 18:00 we prepared a master plate (LB+C) and corresonding liquid cultures from 6 clones of ''E.coli'' NEB10B k1319021-psB1C3. Liquid cultures and master plate were incubated at 37°C. |
| - | * At 23:30 we prepared liquid cultures from K1319015-pSB3K3 clones #7, #8 and #9 in 5 mL LB-medium. Kanamycin was used as antibiotic and the cultures were incubated at | + | * At 23:30 we prepared liquid cultures from K1319015-pSB3K3 clones #7, #8 and #9 in 5 mL LB-medium. Kanamycin was used as antibiotic and the cultures were incubated at 37°C. Purpose of the cultures was cryo stock preparation and plasmid prep. |
==5th == | ==5th == | ||
| Line 156: | Line 224: | ||
* At 12:00 we prepared liquid cultures from K139010-pSB3K3, K139011-pSB3K3, K139012-pSB3K3, K139013-pSB3K3, K139014-pSB3K3, K139015-pSB3K3 in 5 ml LB-medium each. Kanamycin was used as antibiotic. Purpose for the cultures was the characterization of constituitive expression and an additional plasmid prep of K139013-pSB3K3 and K139014-pSB3K3. | * At 12:00 we prepared liquid cultures from K139010-pSB3K3, K139011-pSB3K3, K139012-pSB3K3, K139013-pSB3K3, K139014-pSB3K3, K139015-pSB3K3 in 5 ml LB-medium each. Kanamycin was used as antibiotic. Purpose for the cultures was the characterization of constituitive expression and an additional plasmid prep of K139013-pSB3K3 and K139014-pSB3K3. | ||
| - | * Production of sensor-chips was done accordingly to the [https://2014.igem.org/Team:Aachen/Notebook/Protocols | + | * Production of sensor-chips was done accordingly to the [https://2014.igem.org/Team:Aachen/Notebook/Protocols/detection sensor-chip manufacturing protocol]: |
** At 13:00 we prepared sensor chips from shake flask pre-cultures of BL21 pSB1C3-K1319008+pSB3K3-K1319014, BL21 pSB1C3-K1319008+pSB3K3-K1319013 and NEB pSB1C3-B0015. | ** At 13:00 we prepared sensor chips from shake flask pre-cultures of BL21 pSB1C3-K1319008+pSB3K3-K1319014, BL21 pSB1C3-K1319008+pSB3K3-K1319013 and NEB pSB1C3-B0015. | ||
| - | **Subsequently to 1 h incubation at | + | **Subsequently to 1 h incubation at 37°C, BL21 pSB1C3-K131900+pSB3K3-K1319014, BL21 pSB1C3-K1319008+pSB3K3-K1319013 and NEB pSB1C3-B0015 were induced with 0.2 µL IPTG. The induced sensor-chips were read out every 30 minutes for 360 minutes in total. K1319013 was induced earlier and thus measurements were taken for 450 min in total. Readout was conducted at 480 nm wavelength. An additional readout was conducted next day at 11:00. |
* At 15:30 we prepared liquid cultures for the characterization of ITPG inducible expression: | * At 15:30 we prepared liquid cultures for the characterization of ITPG inducible expression: | ||
| Line 230: | Line 298: | ||
* made a master plate at 15:30 containing two clones of K1319015 plate from yesterday. | * made a master plate at 15:30 containing two clones of K1319015 plate from yesterday. | ||
| - | * At 22:00 we inocculated two 500 ml flasks containing 50 ml LB-medium with 1 ml pre-culture of K1319017, which had been prepared from a master-plate earlier this day. Chloramphinicol was used as antibiotic. The flasks were incubated at | + | * At 22:00 we inocculated two 500 ml flasks containing 50 ml LB-medium with 1 ml pre-culture of K1319017, which had been prepared from a master-plate earlier this day. Chloramphinicol was used as antibiotic. The flasks were incubated at 37°C. One culture (50 ml) was induced with 25 µg homoserinlacton after an OD of 0.6 was reached and the induced culture as well as the control were used for fluorescence characterization. Fluorescene and OD were monitored once per hour. The OD of the induced culture stagnated at ~0.7 while the non induced culture grew further. |
== 7th == | == 7th == | ||
| Line 275: | Line 343: | ||
</center> | </center> | ||
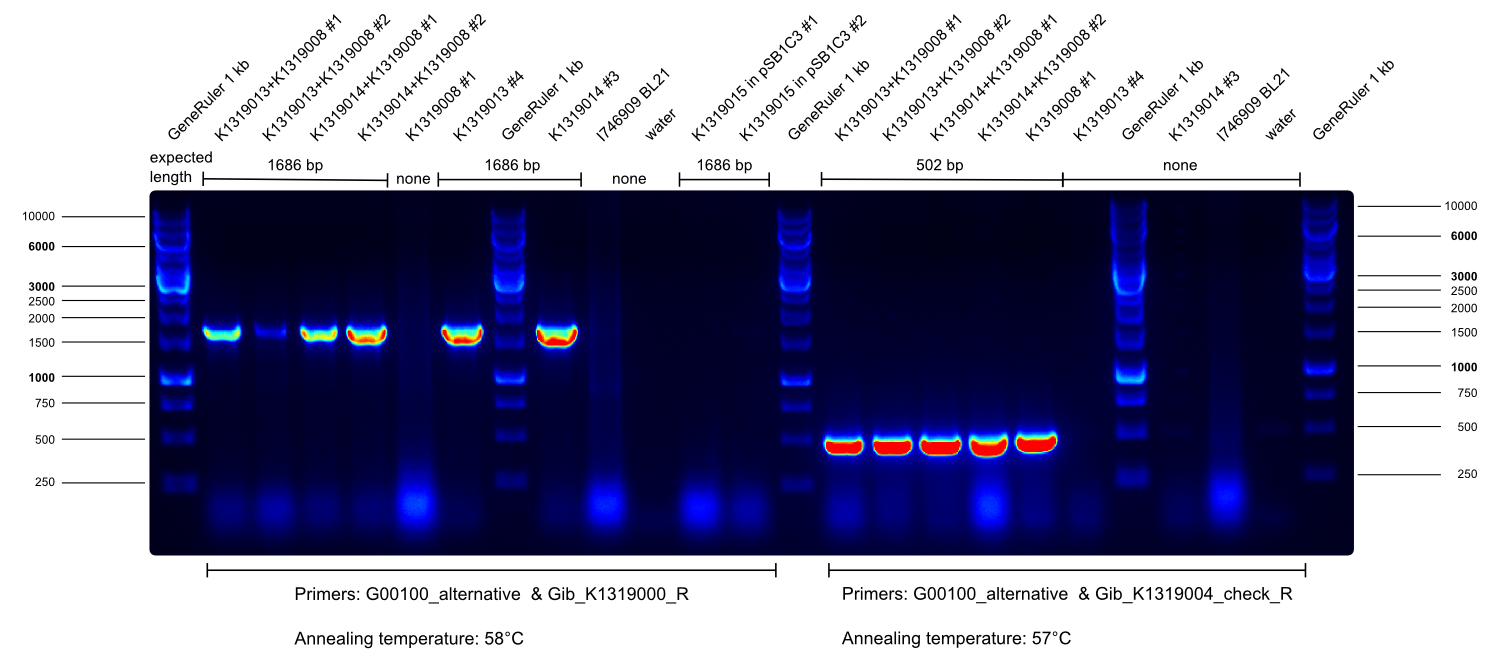
| - | * overnight culture of K1319013 + K1319008 # 1 and # 2, K1319014 + K1319008 # 1 and # 2 for chip production | + | * overnight culture of K1319013 + K1319008 # 1 and # 2, K1319014 + K1319008 # 1 and # 2 for chip production ([https://2014.igem.org/Team:Aachen/Notebook/Protocols/detection sensor-chip manufacturing protocol]). |
== 8th == | == 8th == | ||
| - | * | + | * We prepared sensor-chips with K1319013 + K1319008 # 1 and # 2, K1319014 + K1319008 # 1 and # 2 and B0015 accordingly to the [https://2014.igem.org/Team:Aachen/Notebook/Protocols/detection sensor-chip manufacturing protocol]. Images were made every 30 min with our own device. |
* made precultures with K1319013 + K1319008 # 1 and # 2, K1319014 + K1319008 # 1 and # 2 and K731520. | * made precultures with K1319013 + K1319008 # 1 and # 2, K1319014 + K1319008 # 1 and # 2 and K731520. | ||
* cutting K1319013, K1319014 ans a pSB1A3 vector backbone with EcoRI and SpeI and K1319008 with XbaI and PstI | * cutting K1319013, K1319014 ans a pSB1A3 vector backbone with EcoRI and SpeI and K1319008 with XbaI and PstI | ||
| Line 287: | Line 355: | ||
* SOC medium was added to the transformation before the heat shock had occurred by mistake. | * SOC medium was added to the transformation before the heat shock had occurred by mistake. | ||
* At 02:00 the precultures for the characterization experiment were transferred to 250 ml shake flasks (3 ml culture + 10 ml LB+antibiotics). | * At 02:00 the precultures for the characterization experiment were transferred to 250 ml shake flasks (3 ml culture + 10 ml LB+antibiotics). | ||
| - | * new transformation of the ligation product was conducted into BL21 and | + | * new transformation of the ligation product was conducted into BL21 and DH5α |
* At 23:00 a 500 ml flask containing 50 ml LB-medium was inocculated from a K131026 BL21 cryo stock for sensor-chip manufacturing the following day. | * At 23:00 a 500 ml flask containing 50 ml LB-medium was inocculated from a K131026 BL21 cryo stock for sensor-chip manufacturing the following day. | ||
== 10th == | == 10th == | ||
* made master plates of the new transformation of the ligation product and overnight cultures | * made master plates of the new transformation of the ligation product and overnight cultures | ||
| - | * made chips with K131026 in BL21. Images were taken automaticly every 5 min with our own device | + | * made chips with K131026 in BL21. Images were taken automaticly every 5 min with our own device |
| - | + | ||
| - | + | ||
| - | + | ||
| + | * For sensor-chip manufacturing the following day, we inocculated 500 ml flasks containing 50 ml LB-medium with NEB10β K1319017.pSB1C3 clone #2 and with BL21 K1319042.pSB1C3 clone 2# accordingly to the [https://2014.igem.org/Team:Aachen/Notebook/Protocols/detection sensor-chip manufacturing protocol]. Inoccultion was done at 21:35. | ||
| + | *In preparation for an additional main charcterization experiment we inocculated 200 ml flasks or 300 ml flasks as available containing 25 ml LB-medium with seven of our own constructs listd below. Inoccuaation was done at 22:00. | ||
| + | **K1319010 | ||
| + | **K1319011 | ||
| + | **B0015 | ||
| + | **I20260 | ||
| + | **K1319015 | ||
| + | **K1319014 | ||
| + | **K1319012 | ||
| + | **k1319013 | ||
| + | |||
| + | ==11th== | ||
| + | |||
| + | *At 21:00 we inocculted two 500 ml flasks containing 50 ml LB-medium with BL21 K1319042.pSB1C3 (from plate) and DH5α K1319026.pSB1C3 (from cryo), respectively. Chloramphenicol was used as antibiotik. Both cultures were required for chip manufacturing the next day ([https://2014.igem.org/Team:Aachen/Notebook/Protocols/detection sensor-chip manufacturing protocol]). The chips´ fluorescence was meant to be measured with a plate reader starting ~11:00 the next day. The chips were meant to be measured in parallel. | ||
| + | |||
| + | *At 20:30 eight cultures listed below were plated out for additional characterization experiments on monday. | ||
| + | **B0015 in pSB1C3 | ||
| + | **I20260 in pSB3K3 | ||
| + | **K731520 pSB1C3 #2 | ||
| + | **I746909 in pSB1C3 | ||
| + | **K1319042 i pSB1C3 #2 | ||
| + | **K1319008 in pSB1C3 #1 | ||
| + | **K1319008/13 in pSB1C3/3K3 #1 | ||
| + | **K1319008/14 in pSB1C3/3K3 #2 | ||
| + | |||
| + | ==12th== | ||
| + | |||
| + | *At 9:00 sensor-chips were prepared from precultures of BL21 K1319042.pSB1C3 and DH5α K1319026pSB1C.pSB1C3 accordingly to the [https://2014.igem.org/Team:Aachen/Notebook/Protocols/detection sensor-chip manufacturing protocol]. The cultures were induced at ~11:00 and fluorescence was measured using a plate-reader. K1319026 was additionally measured in our device for its fluorescence. | ||
| + | * Media with different antibiotics for the experiment for collaboration with Heidelberg were prepared. Also media for Robolektor were made. | ||
| + | * Cultures of I13507 in seven vectors listed below were solued in 0,9% NaCl for the OD measurment. Then all cultures and an additional positive control were inoculated in 96-wellsplate with resulted OD of 0,648. The cultures were required for calibration of the plate-reader used for fluorescence measurement, which was part of the Heidelberg chracaterization project. Inocculation was done at ~19:15. | ||
| + | **pSBX1A3 | ||
| + | **pSBX4A5 | ||
| + | **pSBX1C3 | ||
| + | **pSBX4C5 | ||
| + | **pSBX1K3 | ||
| + | **pSBX4K5 | ||
| + | **pSBX1T3 | ||
| + | |||
| + | *At 22:15 a 300 ml flask containing 30 ml LB-medium was inocculated with I746909.pSB1C3. The culture was required for calibration of the robolector (Jülich FHZ). Cloramphenicol was used as antibiotic. | ||
| + | |||
| + | ==13th== | ||
| + | * overnight culture of K131026 in BL21 for chips | ||
| + | |||
| + | ==14th== | ||
| + | * made chips of K131026 in BL21 in LB. Images were taken nearly every 5 or 2 min with our own device. Chips were induced with 2 µl of ''Pseudomonas aeruginosa'' liquid culture or 500 µg/ml HSL | ||
| + | * overnight cultures of K1319013 + 8 and K1319014 + 8 in BL21 for chips | ||
| + | |||
| + | == 15th == | ||
| + | * made chips of K1319013 + 8 and K1319014 + 8 (BL21) in LB. Images were taken every 2 min with our own device and every 4 min in the plate reader. Chips were induced with 2 µl of IPTG (100 mM). | ||
| + | * made overnight cultures of K1319013 + 8, K1319014 + 8, K731520 and I746909 for chips | ||
| + | |||
| + | == 16th == | ||
| + | * made chips of K1319013 + 8, K1319014 + 8, K731520 and I746909 in LB. Images were taken every 2 min with our own device from K731520 and I746909 and every 4 min from all strains in the plate reader. Chips were induced with 2 µl of IPTG (100 mM). | ||
| + | |||
| + | <html> | ||
<center> | <center> | ||
| - | + | <ul class="menusmall-grid"> | |
| - | + | ||
| - | + | <!-- <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | |
| + | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/March" style="color:black"> | ||
| + | <div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to March</div></div> | ||
| + | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/7/7a/Aachen_14-10-10_March_iFG.png); norepeat scroll 0% 0% transparent; background-size:100%; height:100px; width: 100px;"> | ||
| + | </div> | ||
| + | </a> | ||
| + | </li> --> | ||
| + | |||
| + | <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | ||
| + | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/April" style="color:black"> | ||
| + | <div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to April</div></div> | ||
| + | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/2/2d/Aachen_14-10-10_April_iFG.png); norepeat scroll 0% 0% transparent; background-size:100%; height:100px; width: 100px;"> | ||
| + | </div> | ||
| + | </a> | ||
| + | </li> | ||
| + | |||
| + | <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | ||
| + | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/May" style="color:black"> | ||
| + | <div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to May</div></div> | ||
| + | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/6/67/Aachen_14-10-10_May_iFG.png); norepeat scroll 0% 0% transparent; background-size:100%; height:100px; width: 100px;"> | ||
| + | </div> | ||
| + | </a> | ||
| + | </li> | ||
| + | |||
| + | <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | ||
| + | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/June" style="color:black"> | ||
| + | <div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to June</div></div> | ||
| + | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/1/1d/Aachen_14-10-10_June_iFG.png); norepeat scroll 0% 0% transparent; background-size:100%; height:100px; width: 100px;"> | ||
| + | </div> | ||
| + | </a> | ||
| + | </li> | ||
| + | |||
| + | <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | ||
| + | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/July" style="color:black"> | ||
| + | <div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to July</div></div> | ||
| + | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/1/19/Aachen_14-10-10_July_iFG.png); norepeat scroll 0% 0% transparent; background-size:100%; height:100px; width: 100px;"> | ||
| + | </div> | ||
| + | </a> | ||
| + | </li> | ||
| + | |||
| + | <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | ||
| + | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/August" style="color:black"> | ||
| + | <div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to August</div></div> | ||
| + | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/4/4e/Aachen_14-10-10_August_iFG.png); norepeat scroll 0% 0% transparent; background-size:100%; height:100px; width: 100px;"> | ||
| + | </div> | ||
| + | </a> | ||
| + | </li> | ||
| + | |||
| + | <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | ||
| + | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/September" style="color:black"> | ||
| + | <div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to September</div></div> | ||
| + | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/d/d4/Aachen_14-10-10_September_iFG.png); norepeat scroll 0% 0% transparent; background-size:100%; height:100px; width: 100px;"> | ||
| + | </div> | ||
| + | </a> | ||
| + | </li> | ||
| + | |||
| + | <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | ||
| + | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/October" style="color:black"> | ||
| + | <div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to October</div></div> | ||
| + | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/6/60/Aachen_14-10-10_October_iFG.png); norepeat scroll 0% 0% transparent; background-size:100%; height:100px; width: 100px;"> | ||
| + | </div> | ||
| + | </a> | ||
| + | </li> | ||
| + | |||
| + | </ul> | ||
</center> | </center> | ||
| + | </html> | ||
{{Team:Aachen/Footer}} | {{Team:Aachen/Footer}} | ||
Latest revision as of 22:09, 17 October 2014
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"