Team:UC Davis/Potentiostat Design
From 2014.igem.org

Inspiration & Iteration
Inspiration & Iteration
Software
Software & Downloads
Get Started!
Get Started!

The bias encourages diffusion but more importantly the transfer of electrons which are recorded, and ultimately related to the species present in solution. We were faced with the decision to buy or build.
Unfortunately, publication quality potentiostats can range in the tens of thousands of dollars. We needed a cheaper solution if we wanted our device to be consumer friendly. We researched the literature for potentiostat circuits, and were presented with the CheapStat.
Inspiration and Iteration
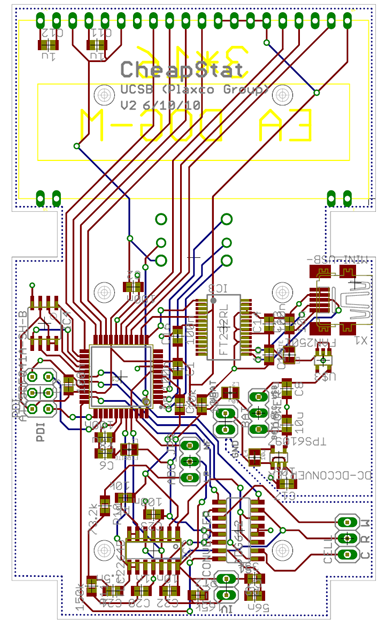 The CheapStat is a budget friendly device developed at UC Santa Barbara several years ago. We aspired to build this circuit and modify the software for our purposes, however, the microcontroller on the cheapStat was controlled predominantly in machine level code. The learning curve seemed unreasonable considering our deadline, so we had to find another path.
The CheapStat is a budget friendly device developed at UC Santa Barbara several years ago. We aspired to build this circuit and modify the software for our purposes, however, the microcontroller on the cheapStat was controlled predominantly in machine level code. The learning curve seemed unreasonable considering our deadline, so we had to find another path.
We were ultimately forced to build our own. With a clean slate, we wanted to create a device that would fulfill our needs, but also be welcomed by the iGEM community. We aimed to match the performance of the cheapStat, but also improve in three ways: increase the effective range of the instrument, decrease the cost of the circuit, and convert to an arduino-friendly microcontroller.
We modeled our circuit after the cheapStat, eventually including more than 30 components. We went on to create five iterative prototypes. The schematic and board files are all available here. To read more about our inspiration and circuit design, click here.
Build
Once we had identified the aldehyde dehydrogenases we wanted to screen for specificities, we needed order DNA so we could express the enzymes and assay them in the lab. For enzymes identified in the literature, sequences were pulled from UniProt Knowledge Base. We ordered DNA from Life Technologies, cloned the genes into the pET29b-(+) plasmid vector using the Gibson assembly method, and transformed the assembled plasmids into BLR strain E. coli for expression. For our engineered mutants, Kunkel mutagenesis was used to introduce desired mutations into the plasmid DNA. Expressed aldehyde dehydrogenase enzymes were purified using affinity chromatography.
Click on “Build” to learn more about how we turned DNA sequences into enzymes
Test
To determine the specificity profiles of our aldehyde dehydrogenases, we needed to develop a simple, high-throughput assay which we could ultimately use to determine the aldehyde composition of a sample of olive oil. We developed a simple spectrophotometric plate assay which measures the concentration of NADH in a solution. Using this assay, we screened all 26 aldehyde dehydrogenases against 16 aldehyde substrates and identified four enzymes with unique specificities. We created full Michaelis-Menten curves and calculated the kinetic constants for the four enyzmes we identified on all sixteen aldehyde substrates. We also developed a protocol for extracting aldehydes from olive oil which could be used with our plate assay.
Click on “Test” to learn more about the results of our assays
 "
"