Team:CU-Boulder/Notebook/Measurement/Notes
From 2014.igem.org
7/1/14 Dry Lab only: Planning and research into this study. From the description on iGEM’s website (https://2014.igem.org/Tracks/Measurement/Interlab_study): The first device is already built and available in the distribution kit. The second and third devices must be built using the BioBricks standard protocol by the teams participating in this study. 1. Existing device: BBa_I20260 (J23101-B0032-E0040-B0015) in the pSB3K3 vector. Kit location Plate 4, Well 18A 2. New device to be built by the iGEM team: BBa_J23101 + BBa_E0240 (B0032-E0040-B0015), must be built in the pSB1C3 backbone Kit locations BBa_J23101 (called BBa_K823005 when in pSB1C3): Plate 1, Well 20K BBa_E0240 (in pSB1C3): Plate 2, Well 24B 3. New device to be built by the iGEM team: BBa_J23115 + BBa_E0240 (B0032-E0040-B0015), must be built in the pSB1C3 backbone Kit locations BBa_J23115 (called BBa_K823012 when in pSB1C3): Plate 1, Well 22I BBa_E0240 (in pSB1C3): Plate 2, Well 24B
PLEASE NOTE: The J23115 part in this year's distribution kit has two mismatched basepairs and instead matches the K823012 sequence (which was already known to have these mismatches). You can either (a) use the J23115 part as distributed or (b) re-clone J23115 and correct the mismatch. Please let us know which path you will choose at measurement at igem dot org. Thank you! (*This note was not originally on the webpage – it was added around July 12th)
7/3/14
Resuspended DNA from 2014 distribution kit to obtain the 4 parts/devices needed for this study Transformation of Device 1 (I20260) into BWF+ comp cells - Two reactions were performed according to transformation protocol, one is negative control - Heat shock was done for 45 sec - Cultures were plated at full strength and 1:10 dilution with 20 ?L each
7/7/14
Examined plates and observed that growth occurred on both, including the negative control plate. This reveals a problem. It was later discovered that the BW comp cells used in this transformation have Kan resistance, thus explaining the growth on the negative control plate. Transformation will need to be repeated using different comp cells; DH5 alpha cells have been suggested and are available. - Notation: K will be used for Kan plates and C for Chloroamp.
7/10/14
Transformation of all the parts needed from the distribution kit was performed. Each well in the distribution kit from iGEM contains 2-3 ng of DNA. So each transformation contains 200-300 pg/?L. All reactions were done following standard transformation protocol including 45 sec heat shock at 42°C. DH5 alpha comp cells were obtained from Dowell Lab for the reaction.
Sample Volume DNA Volume Comp Cells I20260 (Dev1) 1.5 ?L 40 ?L J23101 1.5 ?L 40 ?L J23115 1.5 ?L 40 ?L E0240 1.5 ?L 40 ?L Neg Control (1.5 ?L MQ H2O) 40 ?L
Following the transformation, 25 ?L of mixture plated full strength on ? of plate, 20 ?L at 1:10 dilution plated on other ? of plate containing the appropriate antibiotic.
7/14/14
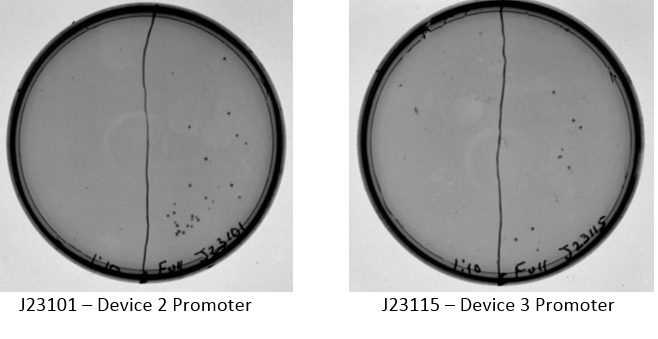
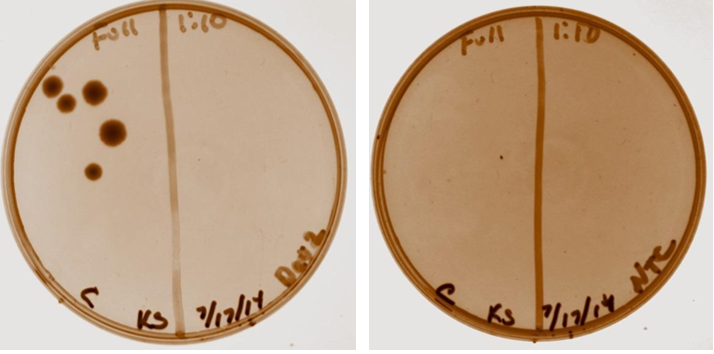
Examination of plates from transformation shows various single colonies in the full strength and very few, if any colonies on the 1:10 dilution side of plates. This may indicate low transformation efficiency or could be due to slow growth of comp cells, which was observed by other iGEM students. Plates images shown below. Colonies were taken from I20260, E0240, and J23101 plates for overnight cultures in the appropriate antibiotic.
I20260 – Device 1 E0240 - GFP
J23101 – Device 2 Promoter J23115 – Device 3 Promoter

Negative Control Plate
7/15/14
Minipreps were done according to manufacturer’s recommendations for each of the overnight cultures. Bacterial pellets were made with the remainder of the cultures. Nanodrop performed on the miniprepped products as shown below:
| Sample | ng/ ?L | 260/280 | 260/230 |
|---|---|---|---|
| Dev1 (I20260) | 43.9 | 1.93 | 2.13 |
| J23101 | 54.7 | 1.88 | 2.15 |
| E0240 | 13.2 | 1.92 | 1.10 |
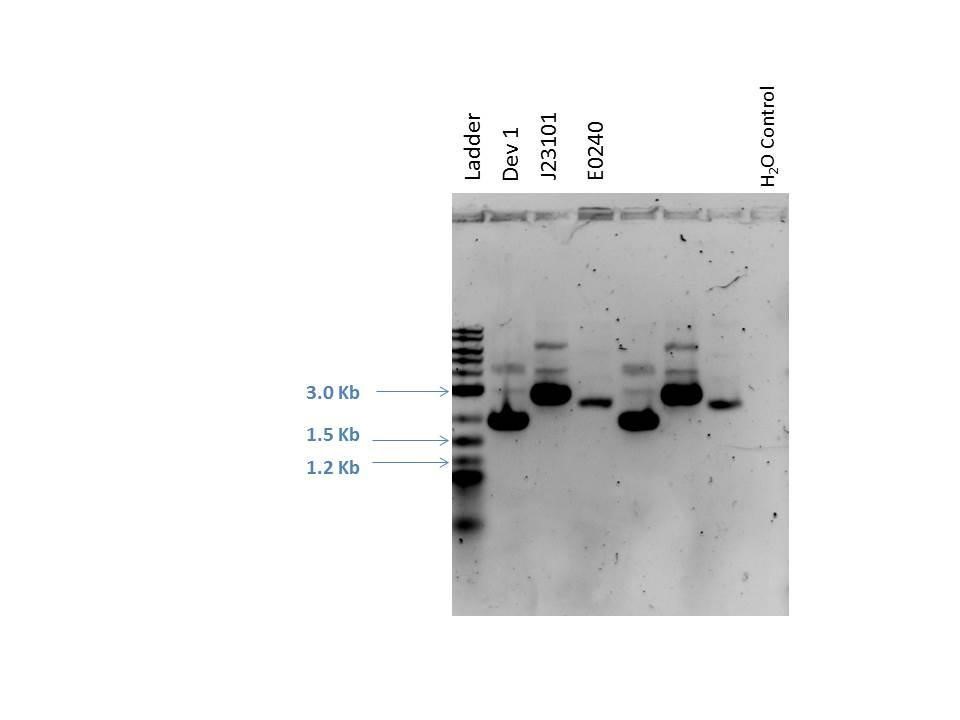
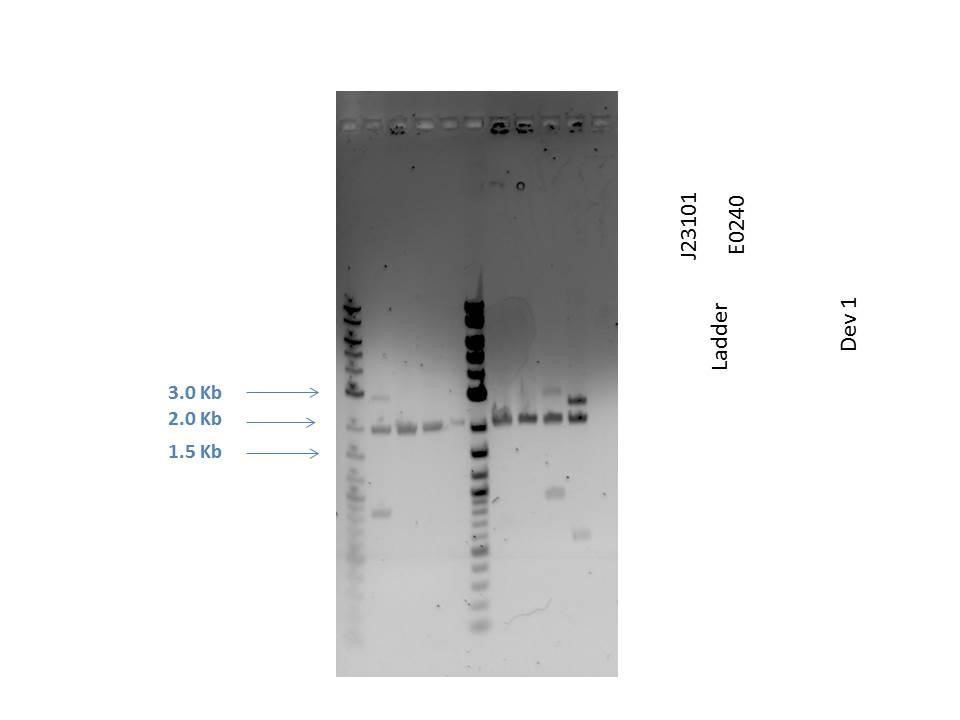
Gel electrophoresis was performed on the products. A 1% agarose gel (0.3 g agarose) with 3.0 ?L EtBr in 30 mL TAE was prepared. 5 ?L of a 2-Log DNA ladder was loaded followed by 5.0 ?L of each sample with 1.0 ?L loading dye into the following lanes:
| Well | Loaded | Expected Size (bp) |
|---|---|---|
| 1 | 2-Log DNA Ladder | N/A |
| 2 | D1 (I20260) | 3669 |
| 3 | J23101 | 2100 |
| 4 | E0240 | 2946 |
| 5 | D1 (I20260) | 3669 |
| 6 | J23101 | 2100 |
| 7 | E0240 | 2946 |
| 8 | H2O Control | -- |
As seen from the gel (shown above), various bands are visible for most of the samples. Since these are uncut plasmids, it is assumed that this is from supercoiling.
7/16/14
Restriction Digestion was performed on the 3 samples, followed by gel electrophoresis to analyze the products isolated. The digestion was done according to protocol using a 1 hour digestion at 37 °C followed by 20 min heat inactivation at 80°C. Enzymes and buffer obtained from NEB.
| I20260 | J23101 | E0240 | |
|---|---|---|---|
| CutSmart Buffer | 5.0 ?L | 5.0 ?L | 5.0 ?L |
| PstI | 1.0 ?L | 1.0 ?L | 1.0 ?L |
| SpeI | -- | 1.0 ?L | -- |
| XbaI | 1.0 ?L | -- | 1.0 ?L |
| DNA (250 ng) | 5.7 ?L | 4.6 ?L | 18.9 ?L |
| MQ H2O | 37.3 ?L | 38.4 ?L | 24.1 ?L |
| Total Volume | 50.0 ?L | 50.0 ?L | 50.0 ?L |
7/17/14
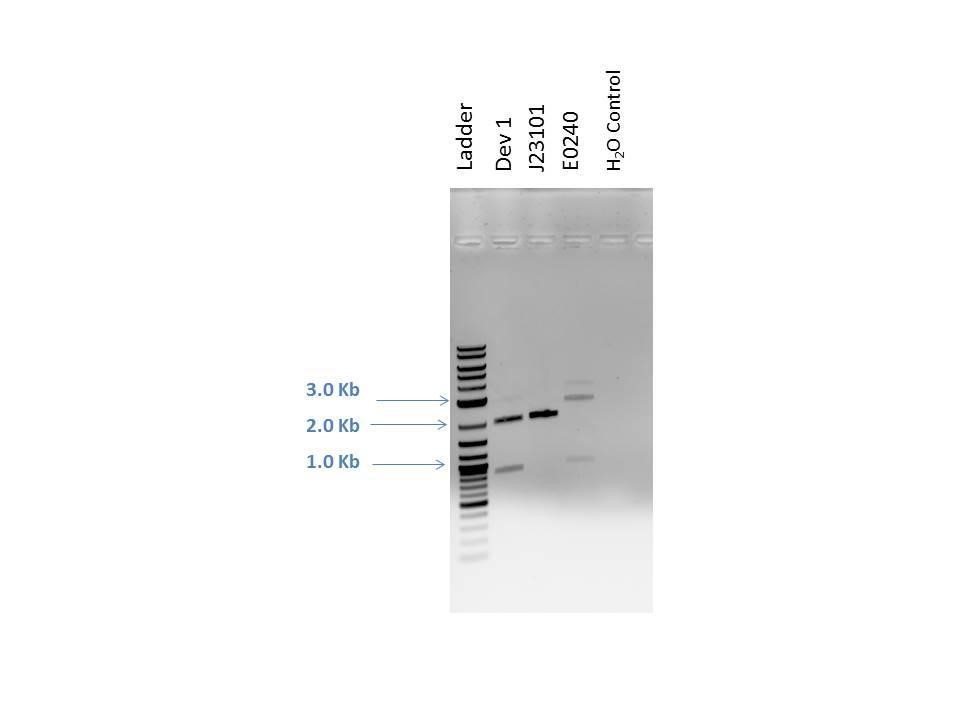
Products from reaction were run on a 1% agarose gel with EtBr. 1.0 ?L of loading dye was added to 5.0 ?L of each sample as well as 5.0 ?L of 2-Log DNA Ladder. Lanes and expected band sizes shown below. There appears to be undigested product present in the E0240 sample.
T4 Ligase 1.25 ?L 1.25 ?L T4 Ligase Buffer 2.5 ?L 2.5 ?L J23101 Vector DNA (40 ng) 8.0 ?L 18.0 ?L E0240 Insert DNA (50 ng) 10.0 ?L -- MQ H2O 3.25 ?L 3.25 ?L Total Volume 25.0 ?L 25.0 ?L
Dev2 #1 124.5 1.85 2.16 Dev2 #2 160.5 1.91 2.29 E0240 #1 150.9 1.82 1.61 E0240 #2 91.0 1.87 2.15 J23115 #1 114.5 1.83 1.97 J23115 #2 148.5 1.79 2.29
| Restriction Digestion was performed on the products using standard protocol and with the following enzymes: |
CutSmart Buffer 5.0 ?L 5.0 ?L 5.0 ?L 5.0 ?L 5.0 ?L 5.0 ?L 5.0 ?L 5.0 ?L PstI 1.0 ?L 1.0 ?L 1.0 ?L 1.0 ?L 1.0 ?L 1.0 ?L 1.0 ?L 1.0 ?L XbaI 1.0 ?L 1.0 ?L 1.0 ?L 1.0 ?L 1.0 ?L 1.0 ?L SpeI -- -- -- -- -- 1.0 ?L 1.0 ?L -- DNA (500 ng) 11.4 ?L 4.0 ?L 3.1 ?L 3.3 ?L 5.5 ?L 4.4 ?L 3.4 ?L -- MQ H2O 31.6 ?L 39.0 ?L 39.9 ?L 39.7 ?L 37.5 ?L 38.6 ?L 39.6 ?L 43.0 ?L Total Volume 50.0 ?L 50.0 ?L 50.0 ?L 50.0 ?L 50.0 ?L 50.0 ?L 50.0 ?L 50.0 ?L
| Ligation reaction was performed to build device 3 from the digested products. This time, several samples will be made. For each reaction, a ratio of 1:3 vector to insert will again be used. A strip of numbered PCR tubes were used for the reactions, with each reaction containing the following reagents. All were placed at 16 °C for 16 hours. |
T4 Ligase Buffer 2.0 ?L J23101 Vector DNA (50 ng) 5.0 ?L E0240 Insert DNA (62.5 ng) 6.25 ?L MQ H2O 5.75 ?L Total Volume 20.0 ?L
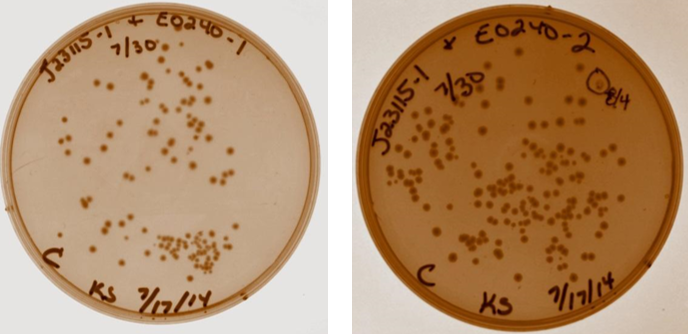
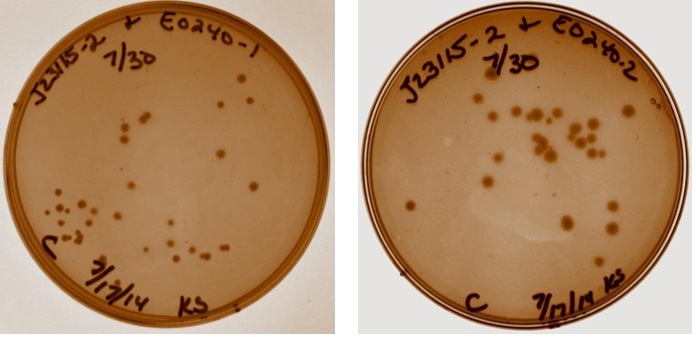
| Sample | Tube # |
| J23115-1 & E0240-1 | 3 |
| J23115-2 & E0240-1 | 4 |
| J23115-1 & E0240-2 | 5 |
| J23115-2 & E0240-2 | 6 |
| Neg Control – J23115-2 and H2O | 7 |
| Empty | 8 |
| 7/30/14 | |
| A heat inactivation was performed on the ligation products. Next, 10 ?L of each of the four device 3 samples and the negative control from tube 7 were transformed into 40 ?L DH5? competent cells (made 7/28). 80 ?L of each was plated after 1.5 hour incubation. | |
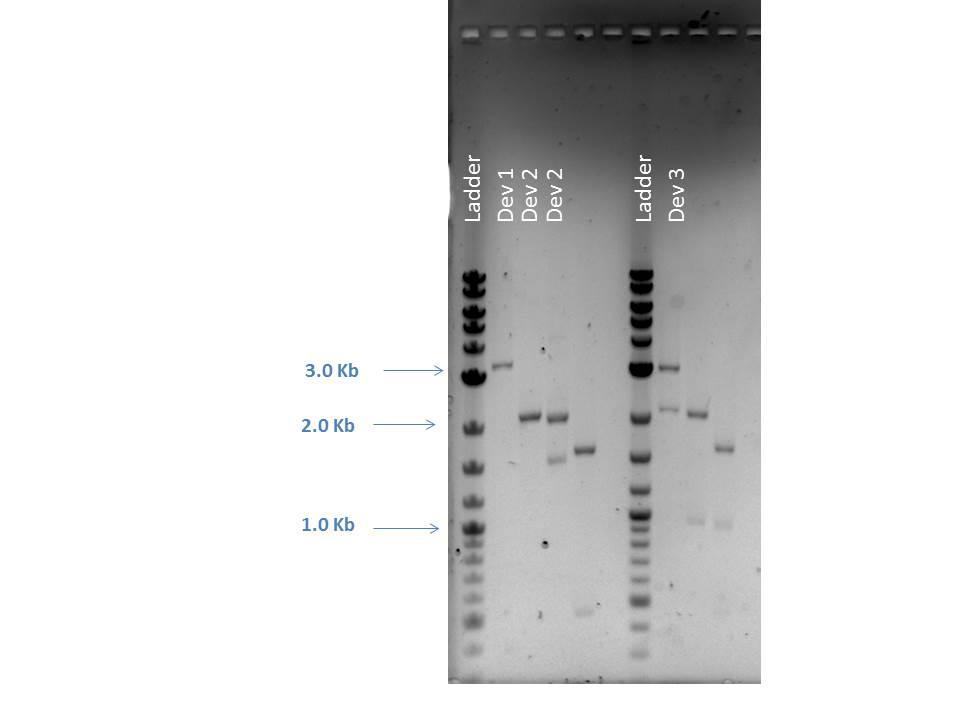
| Gel electrophoresis was performed on digested products from 7/29/14. A 1% agarose gel with EtBr was used and loaded with 5 ?L of ladder and 5 ?L of each sample with 1 ?L loading dye as follows: | |
1 2-Log DNA Ladder N/A 2 D1 2724, 945 3 D2-1 2261 4 D2-2 2042, 900 5 Empty -- 6 2-Log DNA Ladder N/A 7 J23115-1 2100 8 J23115-2 2100 9 E0240-1 2100, 875 10 E0240-2 2100, 875 11 (-) Control --
LB 265 269 259 267 260 Dev 1 1078 541 360 288 249 BL21 780 549 308 212 218
| After subtracting the fluorescence readings by LB alone, both samples demonstrate clear values that decline with decreasing concentrations as expected. The purpose of using the BL21 cell line was to compare cells with known fluorescence against the devices prepared for this study. These results indicate that these cells are producing fluorescence and thus are thought to contain the GFP gene and promoter. With these results, fluorescent measurements can now be taken of all three devices in biological triplicate. After discussing these data with Sam, it was also suggested that cells lacking the GFP gene (such as one of the parts for the promoter for device 2 or 3) be used for a negative control in further studies. These cells would be used to account for fluorescence from the DH5? cells and the pSB1C3 backbone. |
| 8/12/14 |
| Overnight cultures prepared of device 1, 2, and 3. BL21 culture induced with 0.3mM IPTG. |
| 8/13/14 |
| Minipreps and nanodrops prepared from overnight cultures with data shown below. Device 1 and device 2 samples demonstrate very low concentrations. These samples were then used for restriction digestion with multiple enzymes followed by agarose gel electrophoresis (1% agarose with EtBr as previously described). |
Dev 1 9.6 1.90 1.40 Dev 2 35.7 1.87 2.29 Dev 3 230.8 1.87 2.37
| Restriction Digestion |
(200 ng) MQ H2O Total Volume Bands Expected (Bp) D1-Double 2.0 ?L 1.0 ?L -- -- 20.8 ?L 1.16 ?L 25.0 ?L 3296, 373 D2-1 Single 2.0 ?L -- 1.0 ?L -- 5.6 ?L 16.4 ?L 25.0 ?L 2981 D2-1 Double 2.0 ?L -- -- 1.0 ?L 5.6 ?L 16.4 ?L 25.0 ?L 2046, 935 D2-1 Triple 2.0 ?L -- 1.0 ?L 1.0 ?L 5.6 ?L 15.4 ?L 25.0 ?L 1595, 935, 451 D3-1 Single 2.0 ?L -- 1.0 ?L -- 0.86 ?L 21.14 ?L 25.0 ?L 2981 D3-1 Double 2.0 ?L -- -- 1.0 ?L 0.86 ?L 21.14 ?L 25.0 ?L 2046, 935 D3-1 Triple 2.0 ?L -- 1.0 ?L 1.0 ?L 0.86 ?L 20.14 ?L 25.0 ?L 1595, 935, 451
| Well | Loaded | Expected Sizes (bp) |
|---|---|---|
| 1 | 2-Log DNA Ladder | N/A |
| 2 | D1 Double | 3296, 373 |
| 3 | D2 Single | 2981 |
| 4 | D2 Double | 2046, 935 |
| 5 | D2 Triple | 1595, 935, 451 |
| 6 | Empty | N/A |
| 7 | 2-Log DNA Ladder | N/A |
| 8 | D3 Single | 2981 |
| 9 | D3 Double | 2046, 935 |
| 10 | D3 Triple | 1595, 935, 451 |
| 11 | (-) Control | -- |
As seen from the image above, smaller bands ran off the bottom of the gel and could not be readily visualized. However, the approximate sizes of the bands are as expected for each sample. Another 96 well plate was loaded using the sample protocol as previously described. Samples were measured after dilution and regrowth to OD of 0.5. J23101 and BL21 samples were used as controls, raw data shown below.
| Sample | Full | 50% | 25% | 12.5% | 6.25% |
|---|---|---|---|---|---|
| LB | 706 | 268 | 188 | 137 | 123 |
| J23101 | 935 | 294 | 204 | 145 | 125 |
| BL21 | 1277 | 349 | 226 | 159 | 130 |
| Dev 1 | 1623 | 438 | 258 | 182 | 145 |
| Dev 2 | 920 | 303 | 193 | 134 | 123 |
| Dev 3 | 1147 | 367 | 219 | 157 | 126 |
As with the previous test, all 3 devices demonstrate fluorescence in excess of the negative control sample. Values at full and below 12.5% concentration may be beyond the optimal range for the instrument to detect. The dilution amounts should be adjusted for the full test based on these data. 8/18/14
Made LB agar plates with appropriate antibiotics. 100 ?L of each sample was plated and placed in incubator for overnight growth. These will be used to pull individual colonies for the biological triplicates needed in the fluorescent measurements to be performed. A repeat miniprep was performed on the device 1 sample from 8/13/14 in an attempt to obtain a higher concentration.
8/19/14
Plates made yesterday grew too well and single colonies could not be isolated. New streak plates were created for each sample (plates were not imaged).
8/20/14
Three colonies were taken from each streak plate to be used as biological triplicates in the study. Plates are shown below (Top row from left; Dev 1, Dev 2, Dev 3. Bottom row: J23101 negative control, BL21 positive control).
8/21/14
Samples were monitored throughout the day and diluted as necessary to keep them in log phase. Since samples did not grow at the same rate, some difficulties were apparent when trying to get each sample to the same OD at the same time as the other samples. The volume per well will be increased to 150 ?L and the concentrations adjusted for the measurement.
8/22/14
Fluorescence measurements were taken of device 1, 2 and 3 biological triplicate samples as previously described. Samples were all in log phage growth with OD of approximately 0.5 as shown below. Due to the number of samples, two separate 96 well plates were prepared using the same controls. Raw data is shown below.
| Sample | Final OD |
|---|---|
| J23101 | 0.508 |
| Dev 1-1 | 0.515 |
| Dev 1-2 | 0.514 |
| Dev 1-3 | 0.509 |
| Dev 2-1 | 0.490 |
| Dev 2-2 | 0.508 |
| Dev 2-3 | 0.526 |
| Dev 3-1 | 0.512 |
| Dev 3-2 | 0.501 |
| Dev 3-3 | 0.520 |
| 100 | 0.67 | 0.44 | 0.3 | 0.2 | 13.4 | 0.09 | |
|---|---|---|---|---|---|---|---|
| LB Blank | 778 | 785 | 774 | 766 | 780 | 775 | 788 |
| Neg Control (J23101) | 1068 | 903 | 854 | 821 | 807 | 794 | 788 |
| Dev 3 #1 | 1142 | 995 | 895 | 831 | 804 | 809 | 798 |
| Dev 3 #2 | 1125 | 940 | 852 | 823 | 796 | 805 | 788 |
| Dev 3 #3 | 1035 | 951 | 888 | 837 | 824 | 812 | 797 |
| Dev 2 #1 | 992 | 925 | 849 | 812 | 805 | 779 | 779 |
| Dev 2 #2 | 1083 | 924 | 820 | 813 | 791 | 776 | 761 |
| Dev 2 #3 | 1047 | 879 | 824 | 801 | 776 | 782 | 772 |
Plate #1 raw data with sample concentrations shown as decimals.
| 100 | 0.67 | 0.44 | 0.3 | 0.2 | 13.4 | 0.09 | |
|---|---|---|---|---|---|---|---|
| LB Blank | 816 | 825 | 849 | 816 | 844 | 827 | 825 |
| Neg Control (J23101) | 982 | 925 | 886 | 850 | 845 | 833 | 807 |
| Dev 1 #1 | 1728 | 1450 | 1153 | 1024 | 899 | 881 | 838 |
| Dev 1 #2 | 1830 | 1413 | 1142 | 1010 | 921 | 841 | 861 |
| Dev 1 #3 | 1790 | 1446 | 1155 | 975 | 911 | 861 | 854 |
Plate #2 raw data with sample concentrations shown as decimals.
9/4/14
Minipreps done on triplicate samples for J, D1, D2, & D3
9/5/14
Nanodrop done on mini prepped samples (results shown below) followed by restriction digestion using NcoI. The products were run on a 1% agarose gel with EtBr as previously described. This was done to ensure that all of the samples used in the fluorescence measurements are of the correct size. Although the gel is slightly distorted, the size of the bands correspond with the expected products (image not shown).
| Sample | ng/ ?L | 260/280 | 260/230 |
|---|---|---|---|
| Dev 1 #1 | 13.7 | 1.99 | 2.24 |
| Dev 1 #2 | 12.0 | 1.94 | 2.20 |
| Dev 1 #3 | 14.1 | 1.86 | 1.56 |
| Dev 2-1 #1 | 79.0 | 1.87 | 2.24 |
| Dev 2-1 #2 | 68.0 | 1.83 | 2.14 |
| Dev 2-1 #3 | 88.1 | 1.84 | 2.24 |
| Dev 3 #1 | 577.3 | 1.85 | 2.37 |
| Dev 3 #2 | 272.8 | 1.87 | 2.33 |
| Dev 3 #3 | 840.2 | 1.86 | 2.35 |
| J23115 #1 | 254.3 | 1.85 | 1.70 |
| J23115 #2 | 557.1 | 1.82 | 2.26 |
| J23115 #3 | 784.5 | 1.84 | 2.28 |
| Restriction Digestion |
(250 ng) MQ H2O Total Volume Bands Expected (Bp) J1 2.0 ?L 1.0 ?L 1.0 ?L 21.0 ?L 25.0 ?L 1335, 770 J2 2.0 ?L 1.0 ?L 0.9 ?L† 21.1 ?L 25.0 ?L 1335, 770 J3 2.0 ?L 1.0 ?L 0.3 ?L† (0.95 of 1:3) 21.1 ?L 25.0 ?L 1335, 770 D2-1* 2.0 ?L 1.0 ?L 2.5 ?L 14.5 ?L 20.0 ?L 1991, 990 D2-2* 2.0 ?L 1.0 ?L 2.9 ?L 14.1 ?L 20.0 ?L 1991, 990 D2-3* 2.0 ?L 1.0 ?L 2.3 ?L 14.7 ?L 20.0 ?L 1991, 990 D3-1 2.0 ?L 1.0 ?L 0.45 ?L† (0.9 of 1:2) 21.1 ?L 25.0 ?L 1991, 990 D3-2 2.0 ?L 1.0 ?L 0.92 ?L 21.1 ?L 25.0 ?L 1991, 990 D3-3 2.0 ?L 1.0 ?L 0.3 (0.9 of 1:3) ?L† 21.1 ?L 25.0 ?L 1991, 990
| *Digest done with 200 ng and 20 ?L volume 2 hours prior to the other reactions | |
| †Volume after DNA was diluted | |
1 2-Log DNA Ladder N/A 2 J1 (Hole in Gel) 1335, 770 3 J1 1335, 770 4 J2 1335, 770 5 J3 1335, 770 6 2-Log DNA Ladder N/A 7 D2-1* 1991, 990 8 D2-2* 1991, 990 9 D2-3* 1991, 990 10 2-Log DNA Ladder N/A 11 D3-1 1991, 990 12 D3-2 1991, 990 13 D3-3 1991, 990 14 2-Log DNA Ladder N/A 15 H2O Control N/A
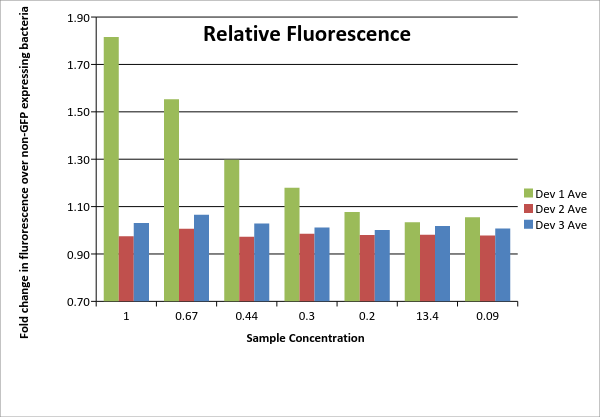
| DATA ANALYSIS |
| Analysis of the fluorescence data measurements obtained is shown below. |
Dev 1 #1 Actual 1728 1450 1153 1024 899 881 838 Calculated 746 525 267 174 54 48 31 Dev 1 #2 Actual 1830 1413 1142 1010 921 841 861 Calculated 848 488 256 160 76 8 54 Dev 1 #3 Actual 1790 1446 1155 975 911 861 854 Calculated 808 521 269 125 66 28 47 Dev 2 #1 Actual 992 925 849 812 805 779 779 Calculated -76 22 -5 -9 -2 -15 -9 Dev 2 #2 Actual 1083 924 820 813 791 776 761 Calculated 15 21 -34 -8 -16 -18 -27 Dev 2 #3 Actual 1047 879 824 801 776 782 772 Calculated -21 -24 -30 -20 -31 -12 -16 Dev 3 #1 Actual 1142 995 895 831 804 809 798 Calculated 74 92 41 10 -3 15 10 Dev 3 #2 Actual 1125 940 852 823 796 805 788 Calculated 57 37 -2 2 -11 11 0 Dev 3 #3 Actual 1035 951 888 837 824 812 797 Calculated -33 48 34 16 17 18 9
| Table 1: Fluorescence data. For each of the three devices, 3 biological replicates were measured (labeled #1, #2, #3 for each device). Data shown includes actual measurements and derived quantities obtained by subtracting fluorescence measured from non-GFP expressing E. coli of the same strain on the same plate. Promoter J23115 for device #3 was used as distributed. In addition, each sample underwent a serial dilution (concentrations listed as fractions across top of table). |
Dev 1 Ave 1.82 1.55 1.30 1.18 1.08 1.03 1.05 Dev 1 Std. Dev 0.05 0.02 0.01 0.03 0.01 0.02 0.01 Dev 2 Ave 0.97 1.01 0.97 0.98 0.98 0.98 0.98 Dev 2 Std. Dev 0.04 0.03 0.02 0.01 0.02 0.00 0.01 Dev 3 Ave 1.03 1.07 1.03 1.01 1.00 1.02 1.01 Dev 3 Std. Dev 0.05 0.03 0.03 0.01 0.02 0.00 0.01
Total: Dev 1 1.23 Total: Dev 2 0.99 Total: Dev 3 1.02
 "
"