Team:CU-Boulder/Notebook/CC9 Team/Constitutive
From 2014.igem.org
Contents |
Week 1
6/4
- Resuspended BBa_K1218011 (biobricked pCas9) from Distribution Kit (4M)
- -Part biobricked by Stanford-Brown 2013
- -Part contains native tracrRNA, promoter, and Cas9 (the Cas9 was modified to remove an EcoRI digestion site) along with a minimal CRISPR array
- Made chemically competent DH3alpha cells
- Tested new competent cells through transformation
- -DNA
- 1ul p11+RBS(2) (4/23)
- 2ul 2B
- 2ul 2P
- 3ul 2P
- No DNA
- -DNA
- -Plated onto Chlor
- -Also, thawed 5a cells for 45 minutes then returned to freezer
- Tomorrow, we will transform and test competency
- Also include non-competent control
6/5
- Transformation results from 6/4
- - No growth on negative (no DNA) control: no contamination
- - > 400 colonies for positive (p11+RBS (2), diluted 1:10) control. Lawn when not diluted
- - ~100 colonies for 2B (2ul) not diluted, 18 colonies on 1:10 dilution
- - 3 colonies for 2P (2ul) not diluted, 0 on 1:10 dilution
- - 20 colonies for 2P (3ul) not diluted, 1 on 1:10 dilution
- Made overnights of 2B and 2P to make freeze downs tomorrow
6/7
- Made 6mL O/N cultures
- -4 from (3) cas9 plate
Week 2
6/8
- Check colonies for presence of Cas9 plasmid
- -Mini-prepped O/Ns from 6/7
- -Digest with EcoRI and PstI (10ul reactions)
- -Run results on a gel
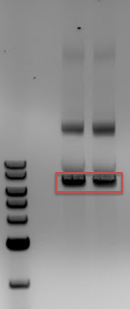
- All 4 samples had the expected bands of 2000 and 5000bp showing that we had Cas9 safely inside cells
- Gel of mini-preps containing Cas9
- -2-log ladder
6/9
- Freeze downs
| Top Label | Side label | What? |
|---|---|---|
| Cas9 6/9 | From dis kit 1 | Cas9 construct from Sanford-Brown team |
| -> K1218011 (tracrRNA, Cas9, minimal CRISPR) | ||
| Cas9 6/9 | From dis kit 2 | “ “ |
- Overnight cultures of Cas9 (Qi wrCas9 #P44250) and gRNA (Qi gRNA #P44251 cells from Andrew
Week 7
7/11
- Trying to get pSB1C3-pCas9 (BBa_K1218011) to function
- -Ordered 2 primers pairs so we could make 2 targeting guide RNAs
- These guide RNAs target the Neocin (also Kanamycin) resistance gene in our BW strain.
- -Ordered 2 primers pairs so we could make 2 targeting guide RNAs
- Vector Digest
- Digest 1-2ul of pCas9 with BsaI-HF
- -Sample was divided between two lanes
- -Gel extracted digestion but poor yield
- -Redigest with less DNA
- 1. pCas9 from above gel
- 2. Different sample of pCas9
- -Repeated, doing everything we could to increase gel extraction yield-- no luck
- Phosphorylation
- -phosphorylated primer pairs together
- -1 hr at 37C then 20min at 65C
- Annealing
- -(forgot to add 2.5ul of 1M NaCl)
- -Incubated in Thermocycler for 5 minutes at 95C
- -Tried to decrement temp 0.1C/s until temp reached 21, then jumped to 4C
- Took less than 30 minutes so we think the samples cooled too fast
7/12
- Trying to switch the gRNA in the CRISPR
- -Phosphorylation
- -Phosphorylated primer pairs same as 7/11
- -Annealing
- -Remembered to add 2.5ul of 1M NaCl
- -1. Heat block
- 5min at 95C-> removed from head to let cool 2 hours
- Problem-> water condenses on lid
- -2. Thermocycler
- Was supposed to cool slowly… not the case
- -Vector Digest
- -Digested pCas9
- 1. 60 min at 37, 20 min at 80 (see #4 on gel below)
- 2. 90 min at 37, 20 min at 80 (see #5 on gel below)
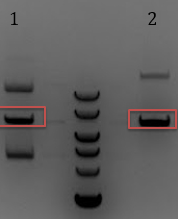
- -Compared to un cut on a gel…. all bands looked pretty. Also, included gel extractions from 7/11 to test whether DNA was present
- -Digested pCas9
- -Phosphorylation
- 1. Gel extract 1 from 7/11
- 2. Gel extrac 2 from 7/11
- 3. Uncut pCas9
- 4. Digested pCas9 for 60 minutes
- 5. Digested pCas9 for 90 minutes
- -Did not gel extract sample
- -Phosphorylation #3
- -Because I did not like my previous annealing attempts, so I decided to start over
- -Annealing
- -Added 2.5ul 1M NaCl
| 95C | 94.8 | 94.6 | 94.4 | 94.2 | 94.0 | 21 |
|---|---|---|---|---|---|---|
| 5 min | :20 | :20 | :20 | :20 | :20 | hold |
| decrement | -1 | -1 | -1 | -1 | -1 |
- -font cycled 74 times. After each step, the temp would decrement by 1 to produce a consistent cooling rate of 0.2C/20s (about equivalent to letting a heat block cool
- -Diluted 1:10
- -Ligation
- -4 reactions total
- gRNA1- gel extracted pCas9
- gRNA1- non-gel extracted pCas9
- gRNA2- gel extracted pCas9
- gRNA2- non-gel extracted pCas9
- -4 reactions total
Week8
7/13
- Transformation of 7/12 ligations (attempts to swap gRNAs in the CRISPR)
- -Plated on low Chlor (34ug/mL) and high Chlor (170ug/mL)
7/14
- Did get colonies for pCas9 with new gRNAs (some for each primer pair)
- -Colony PCR using VR and Fwd gRNA primer
- -Dream taq
- -Gel had big black spot
- -(Realized this the next day that the imager was set to high exposure so could not see bands)
- -Set up O/N of colonies in despair
- -Colony PCR using VR and Fwd gRNA primer
7/15
- Check colonies for pSB1C3-pCas9 using mini-prepped DNA
- -Mini-prepped O/Ns
- -Repeated PCR using VR and gRNA primer
- -Found some samples with new gRNA
- -Tried to make O/Ns of correct samples….. turns out there was a miscommunication and DNA, not living cells, were added to the liquid cultures…
- First proof of concept: Transformation of pSB1C3-pCas9 containing targeting gRNAs into BW23115 chemically competent cells and 5alphas
- -DNA
- pSB1C3-pCas9(unmodified)--> non-targeting
- pSB1C3-pCas9(gRNA1)--> targeting
- pSB1C3-pCas9(gRNA2) --> targeting
- -Controls
- -5alpha cells. These cells do not contain kanamycin resistance in their genome so should not be targeted
- -pSB1C3-pCas9(unmodified): gRNA does not target anywhere in either E. coli genome so is we expect no targeted killing; hence no death
- -Hypothesis: gRNA1 and gRNA2 are complimentary to the kanamycin resistance gene in the BW23115 strain. Therefore, we expect the samples that contain pCas9 with these targeting gRNAs to target and cleave the resistance gene and cause either direct cell death or a loss of function of the kanamycin resistance gene, resulting in cell death.
- -DNA
7/17
- Results of 7/15 pCas9 transformation experiment
- -5alphas
- No growth for No DNA control on Chlor
- Much growth for non-targeting gRNA and targeting gRNA1
- Half as much growth for targeting gRNA2
- -BW23115
- No growth for No DNA control on Chlor+Kan
- Fewer, but larger colonies for non-targeting sample
- More growth for targeting gRNA1 and gRNA2? large range of sizes
- -Conclusions
- Targeting the Kanamycin resistance gene did not kill the bacteria.
- -Repeat the transformation with remaining DNA samples
- -5alphas
7/18
- Made liquid O/Ns of gRNAs to remake pSB1C3-pCas9(gRNA1) and pSB1C3-pCas9(gRNA2)
7/19
- Tested overnights from 7/18 pSB1C3-pCas9 with gRNAs 1 and 2
- -Mini-prepped
- -Repeated PCR with DreamTaq
- used VR (reverse primer for iGEM biobricks (amplifies insert from end)) and fwd primer used to make gRNA
- There should only be a product if the fwd primer can anneal to DNA. Product should be about 400bp
- -Found some samples with correct band at 400bp- these were the correct sizes so are likely correct
Week 9
7/20
- Freeze downs of samples from 7/19 that looked right
- -gRNA1 targets nt 23-53 of KanR gene
- -gRNA2 targets ny 551-582 of KanR gene
7/21
- Sent samples for sequencing
- -pSB1C3-pCas9(gRNA1)
- 5'- cgatagaaggcgatgcgctgcgaatcggga -3'
- -pSB1C3-pCas9(gRNA2)
- 5'- tcgagcacagctgcgcaaggaacgcccgtc -3'
- -pSB1C3-pCas9(gRNA1)
7/22
- Pricked colony from pSB1C3-pCas9 plate
- -Mini-prepped DNA to isolate DNA
7/23
- Sequencing samples from 7/21 were lost in the mail. Resent samples
- Primers came in for new guide RNAs
- -Note: Upon review of the protocol, we realized that the first round of primers we bought were incorrectly made, resulting in a 1nt shift away from the PAM site. The NGG motif requirement was not met so binding was absent. We bought new primers to correct this problem
- -gRNA3: Targets same place as gRNA1 but is corrected to bind the correct distance from the PAM site
- 5'- gatagaaggcgatgcgctgcgaatcgggag -3'
- -gRNA4: Targets same place as gRNA2 but is corrected to bind the correct distance from the PAM site
- 5'- cgagcacagctgcgcaaggaacgcccgtcg -3'
- -unmodified: spacer with the BsaI sites that allow for replacement of spacer
- 5'- tgagaccagtctcggaagctcaaaggtctc -3'
- To swap gRNAs on pCas9 (gRNA3 and gRNA4)
- -Phosphorylate primers 1hr @ 37C, 20 min @ 65C
- -Annealed primers
- Used the thermocycler like on 7/12
- -Digest vector DNA (pCas9 DNA with BsaI)
- -Did not get complete digestion
- -Digest vector DNA (pCas9 DNA with BsaI)
7/24
- To swap gRNAs on pSB1C3-pCas9 (gRNA3 and gRNA4)
- -Used digested pSB1C3-pCas9 from 7/12. Same one used the last time we swapped gRNAs
- -Ligated the above pSB1C3-pCas9 to annealed oligos
7/25
- Transformed ligation from 7/24 (gRNA1/gRNA2 into pSB1C3-pCas9) into 5alpha cells
7/26
- No growth for 7/25 transformation
- -Retried transformation
- -pCas9 was digested a week earlier- possible source of problem
- Retried making pCas9 with gRNA3 and gRNA4
- -Digestion (so we have freshly cut samples)
- pSB1C3-pCas9 with BsaI
- -Digestion (so we have freshly cut samples)
- -gel extracted linearlized pCas9 (smaller band)
- -Ligation (10hrs at 16C, 10min at 80C)
- pSB1C3-pCas9 + gRNA3
- pSB1C3-pCas9 + gRNA4
Week 10
7/27
- Transformation results
- -No growth for no DNA controls--> on Chlor
- -No growth for gRNA4 --> on Chlor
- -3 colonies for gRNA3 --> on Chlor
- Picked the 3 colonies for gRNA3 for O/Ns
7/28
- All O/Ns grew
- To test the overnights for gRNA3
- -Mini-prep samples -> good yields
- -PCR with Dream-Taq
- -Diluted all samples 1:100
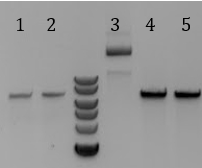
- 1. pSB1C3-pCas9(unmod.)
- 2. Sample 1
- 3. Sample 2
- 4. Sample 3
- 5. No DNA control
- -All samples contained a band of the expected <400bp, including the Stanford-Brown Cas9 unmodified and no DNA control
- Transformations
- -pSB1C3-pCas9(gRNA3)
- -SB1C3-pCas9(gRNA4)
- -Positive control with pSB1C3-pCas9(unmod.)
- -No DNA control
7/30
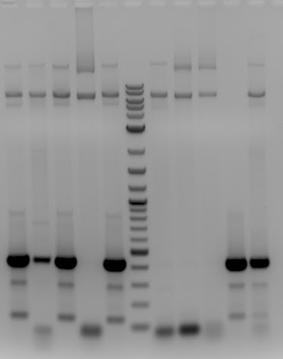
- Sent samples for sequencing
- -pSB1C3(gRNA3) (5) VR
- -pSB1C3(gRNA3) (6) VR
- -pSB1C3(gRNA3) (7) VR
7/31
- Sequencing results came back positive
- -Had correct gRNA3 insert
- Transformation experiments with gNRAs: Repeat of 7/15 experiment
- -Samples
- -Scramble- unmodified pCas9
- -non-targeting- gRNA1
- -targeting- gRNA3
- -no DNA control
- -Added 578ng each
- -Transformed into 5alpha and BW (F+, +KanR)
- -Samples
8/1
- Transformation results from 7/31
- -Contamination for 5alpha samples
- -BW cells on Chlor
- -Hundreds of colonies for all (unmodified, gRNA1, gRNA3)
- -No colonies on no DNA control
- -BW cells on Chlor + Kan
- -Hundreds of colonies for unmodified and gRNA1
- -3-4 colonies for gRNA3
- -No colonies for no DNA control
- -Notes:
- Unexpected that for gRNA3, cells died on Chlor+Kan but survived at comparable amounts on Chlor
- -Suggests that targeting of an essential gene
- -Later, cells on gRNA3 plate looked sickly, maybe delayed killing?
Week 11
8/4
- Spotted contamination in SOC-> remade SOC
- Got new 5alpha (from NEB) from Mike to make comp cells (C2987 by NEB)
- Transformation experiment (~578ng)
- -(barrier tips, new SOC, BW cells (5/30))
- -unmodified pCas9
- -gRNA1
- -gRNA3
- -No DNA control
- Test pCas9 for functional Cas9 rather than dCas9
- -Digestion to confirm
- -pCas9 EcoRV no extraction- just image on gel
- To add gRNA4 to pSB1C3-pCas9
- -Digestion
- pCas9 BsaI extracted entire plasmid
- insert gel
- -Ligation (10 hrs at 16C, 10 min at 80C)
- pSB1C3-pCas9 + gRNA4
- No DNA control to test for contamination in reagents
- -Digestion
8/5
- Transformation into ER2738 (7/18), 5ul of ligation products
- 1. Litmus28ibb-pCas9gRNA3
- 2. pSB1C3-Cas9 (gRNA4)
- 3. No DNA in ligation mix
- No DNA control
- Variations on protocol
- -Added 200ul SOC to 1.7ul epi-tubes, incubated in small tubes
- -Incubated at ~200rpm
- -Added only 5ul Ligation products
- Make chem comp 5alpha cells
- -Got new 5alpha cells from Mike (chem comp from NEB) on 8/4
- -grew O/N in 5mL LB
- -Made fresh .8M MgCl2 and .2M CaCl2
- Transformation results from 8/4 [not targeted killing experiment]
- -No growth when only SOC was plate
- SOC is not contaminated with cells ChlorR or ChlorR+kanR
- -No growth for No DNA controls on A+T or C+T
- Negative Ligation control
- Negative transformation control
- -No growth for M13g-3C5 or LCR ligations (8/3)
- -Lawn for amilCP on pSB1C3 transformation
- -No growth when only SOC was plate
- Targeted killing experiment Transformation results
- -Images of plates can be found in the results section
- -Colony count on chlor (34ug/mL) and kan (25ug/mL) - kanR is essential
| 1:1 | 1:10 | |
|---|---|---|
| unmodified | lawn | 2760 |
| gRNA1 | lawn | 2316 |
| gRNA3 | 23 | 1 |
| no DNA control | 0 | NA |
- Colony count on chlor (170ug/mL) - kanR is non-essential
| 1:1 | 1:10 | |
|---|---|---|
| unmodified | 1920 | 226 |
| gRNA1 | 960 | 71 |
| gRNA3 | 8 | 1 |
| no DNA control | 0 | NA |
- -Shows successful killing when KanR is essential and when KanR is nonessential
8/6
- Transformation results from 8/5
- -No colonies for pCas9(gRNA4) on Tet+reduced Chlor
- -BLASTed sequence against MG1655. The 10nts at the 3’ end of spacer match a sequence in MG1655 that is adjacet to a PAM site. We think gRNA is targeting the production strain.
- To compare, the spacer of gRNA3 matches 8nts on its 3’ end.
- -BLASTed sequence against MG1655. The 10nts at the 3’ end of spacer match a sequence in MG1655 that is adjacet to a PAM site. We think gRNA is targeting the production strain.
- -No growth for no DNA control of ligation or transformation
- -Ligation reactants are not cause of recent contamination on Amp or Chlor
- -No colonies for pCas9(gRNA4) on Tet+reduced Chlor
Week 13/14
8/12
- Targeted Killing Transformation
- -Transform into 3alpha (7/28) and BWF’ (5/30)
- -DNA
- pSB1C3-pCas9(unmod.)
- pSB1C3-pCas9(gRNA1)
- pSB1C3-pCas9(gRNA3)
8/16 Results of 8/12 transformation of BWF’ with gRNA(unmod, 1, or 3) targeted killing
No growth on Chlor (34ug/mL) or Chlor (34ug/mL)+Kan(25ug/mL)
| Chlor+Kan | Chor
| |
|---|---|---|
| Scramble | Healthy lawn | Healthy lawn |
| gRNA1 | Healthy lawn (thinner that scramble) | Healthy lawn (thinner that scramble) |
| gRNA3 | Shadowy, flat lawn, 42 healthy colonies | Shadowy, flat lawn, 42 healthy colonies |
8/19
Sequence survivor colonies from transformation experiments. These are the colonies from gRNA3 samples.
 "
"