Team:ETH Zurich/project/infopro
From 2014.igem.org
Information Processing
Engineering ways to make biological systems perform computation is one of the core goals of synthetic biology. We generally work at the DNA level, engineering systems to function using BioBricks. In most biological systems, protein-protein interactions are where the majority of processing takes place. We design proteins to accomplish computation would allow for systems to function on a much faster timescale than the current transcription-translation paradigm. These are some of the challenges that face teams entering projects into the Information Processing track in iGEM.
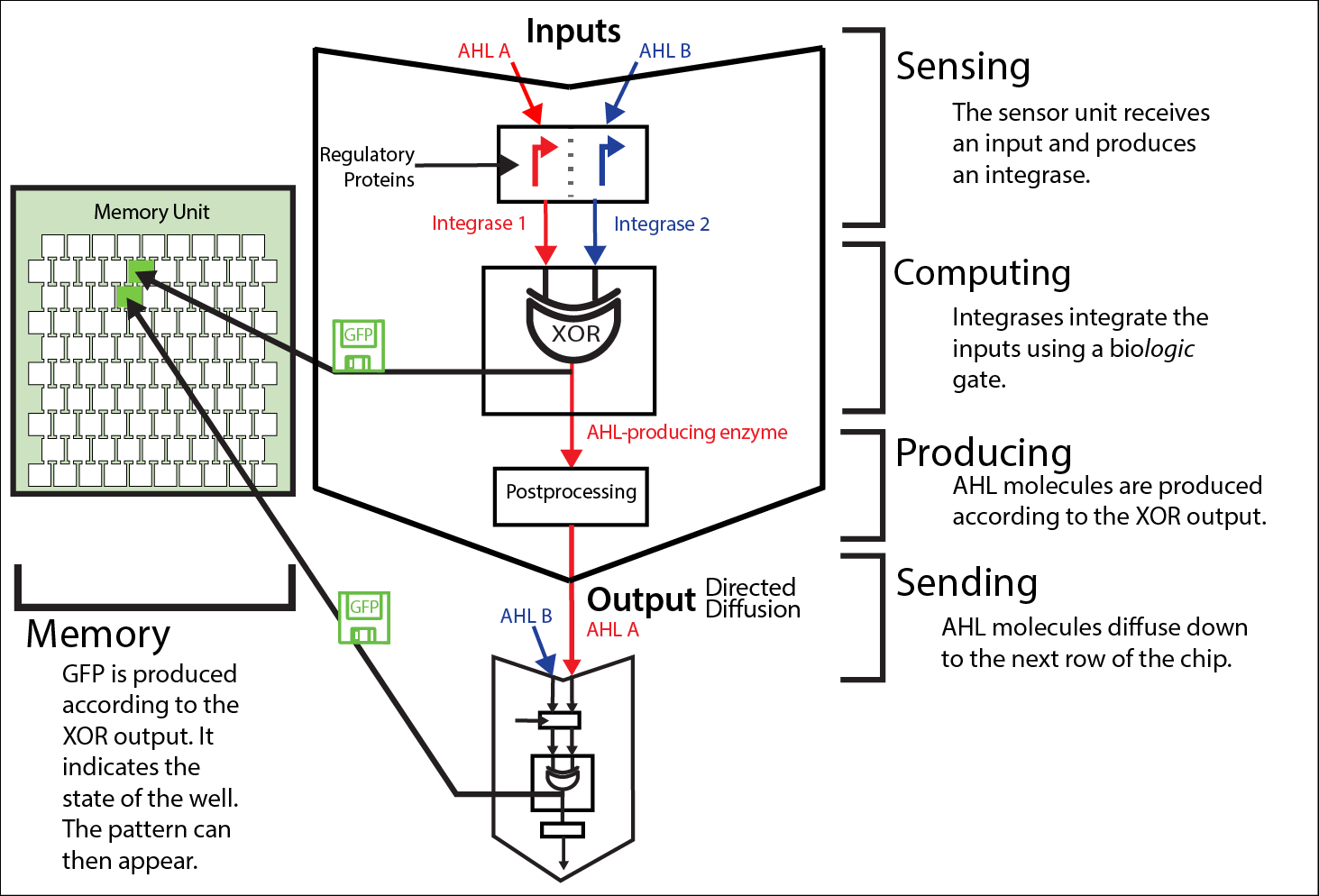
Sensing
Diffused HSL molecules are identified via quorum sensing. This integration from a continuous variable, the concentration of HSL, to a discrete variable, the activation of a promoter, is prone to errors due to leakiness and crosstalk between HSL molecules, regulatory proteins and promoters (we characterized it for [http://parts.igem.org/Part:BBa_R0062 Lux promoter (BBa_R0062)], [http://parts.igem.org/Part:BBa_R0079 Las promoter (BBa_R0079)] and [http://parts.igem.org/Part:BBa_R0071 Rhl promoter (BBa_R0071)]). To limit the risk of error propagation throughout the rows, we used a [http://parts.igem.org/Part:BBa_K1541000 riboregulating system].
Computing
Integrases compute an XOR logic gate via single switching or double switching of a terminator. Their behavior depends on a set of parameters.
Producing
The enzyme synthesizes the signaling molecule AHL.
Sending
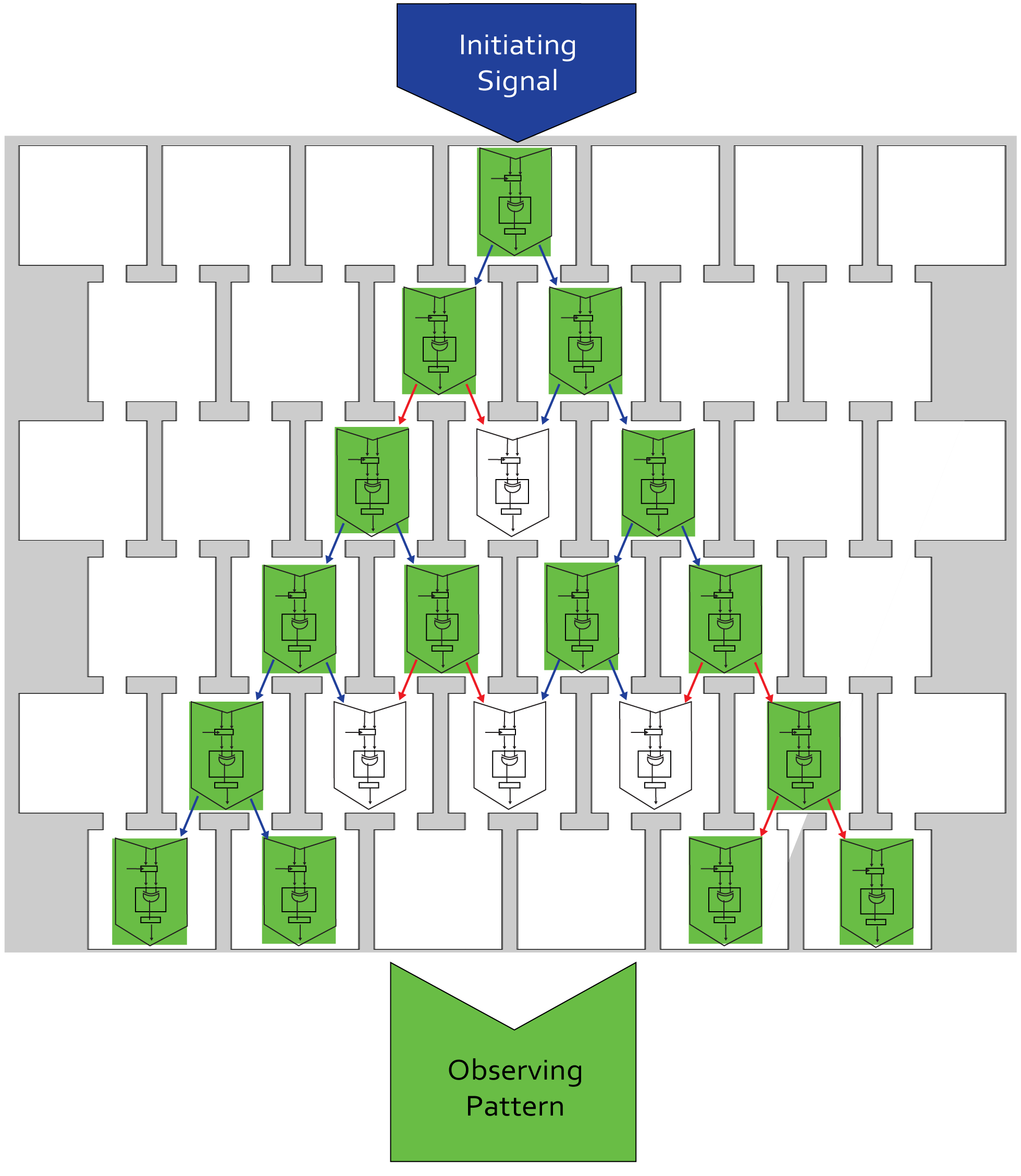
The AHL produced diffuses then through the channels to the rows below. The speed of this directed diffusion determines the time needed from the pattern to emerge. You can find out more information about diffusion and chip design on our modeling page, our experimental results page and our chip page.
Memorising
 "
"