Team:ETH Zurich/project/overview
From 2014.igem.org
(→Implementation in E. coli) |
(→Implementation in E. coli) |
||
| Line 43: | Line 43: | ||
=== Implementation in ''E. coli'' === | === Implementation in ''E. coli'' === | ||
| - | Mosai''coli'' involves three main constructs per cell, one for quorum sensing, one for production of integrases, and the last with the integrase based XOR logic gate to perform computation. Each cell can receive two orthogonal AHLs - the RhlI and LuxI products. The Rhl-AHL or the Lux-AHL received by the cell bind to their corresponding receptor proteins RhlR and LuxR, thus activating them. The LuxR-Lux-AHL or RhlR-Rhl-AHL complex bind to the corresponding promoters Prhl and Plux on the second plasmid and positively regulate the expression of two integrases phiC31 and Bxb1 respectively. Additionally, we use | + | Mosai''coli'' involves three main constructs per cell, one for quorum sensing, one for production of integrases, and the last with the integrase based XOR logic gate to perform computation. Each cell can receive two orthogonal AHLs - the RhlI and LuxI products. The Rhl-AHL or the Lux-AHL received by the cell bind to their corresponding receptor proteins RhlR and LuxR, thus activating them. The LuxR-Lux-AHL or RhlR-Rhl-AHL complex bind to the corresponding promoters Prhl and Plux on the second plasmid and positively regulate the expression of two integrases phiC31 and Bxb1 respectively. Additionally, we use riboregulators to control the expression of the integrases thus reducing the leakiness of promoters Plux or Prhl<sup>[[#refWilliams|[10]]]</sup>. |
[[File:ETHZurich molbio simplified.png |500px|center|]] | [[File:ETHZurich molbio simplified.png |500px|center|]] | ||
Revision as of 06:19, 12 August 2014
{{{1}}}
Summary
Emergence of complex patterns in nature is a fascinating and widely spread phenomenon, which is not fully understood yet. Mosaicoli aims to investigate emergence of complex patterns from a simple rule by engineering a cellular automaton into E. coli bacteria. This automaton comprises a grid of colonies on a 3D-printed millifluidic chip. Each colony is either in an ON or OFF state and updates its state by integrating signals from its neighbors according to a genetically pre-programmed logic rule. Complex patterns such as Sierpinski triangles are visualized by fluorescence after several steps of row-wise propagation. Sequential logic computation based on quorum sensing is challenged by leakiness and crosstalk present in biological systems. Mosaicoli overcomes these issues by exploiting multichannel orthogonal communication, riboregulators and integrase-based XOR logic gates. Engineering such a reliable system not only enables a better understanding of emergent patterns, but also provides novel building blocks for biological computers.
Background
"The whole is more than the sum of its parts" 'Aristotle'
Emergence describes how novel properties can arise at a certain level from lower-complexity parts, where these properties are not observed. [12] This phenomenon is wide-spread in nature. One straight-forward example is water : from the molecules of H2O, who are here considered as simple subparts, we are able to consider the liquid "water". At a higher level of complexity.
One particular pattern formation retained our attention : some sea snail present a complex pattern on their shells. The "Cloth of Gold" observed on textile cones results from an inner computation of the neural cells of the sea snail. This pattern appears row by row (computation after computation) and the pigments of the next row are determined by the pigments of the row before.
This pattern formation was formalized by Neuman in the concept of Cellular Automata. Following a simple pre-programmed logic rule, the state of a new spot, corresponding to the colors (eiter white or brown) in sea snail shells, is determined by the states of three parent spots (from the previous computation round). Wolfram [11] elaborated a whole theory on cellular automata and presented the emergence properties of complex patterns from the wide variety of existing simple rules.
Patterns formed after the cellular automata's theory offer a large panel of properties : striking examples are the rule 30 which gives an apparently random pattern and the rule 110 has been proven to be Turing complete. With cellular automata, you can not predict how the final pattern will look like even if you know the rule that governs its property. Tus, the intricated computations of steps poses the problem of the complexity.
Our project : Mosaicoli
Principle and Goals
The aim of our project is to investigate the emergence of complexity and how we can deal with it. Our project adresses this goal in two ways.
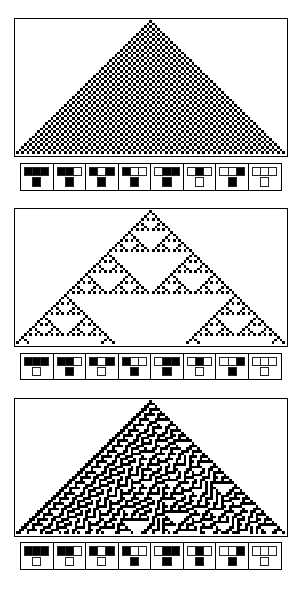
First, we follow a biomimetic approach, and reproduce emergent complex patterns inspired by those that we can observe in nature. This approach corresponds to the moto "What I cannot build, I cannot understand." We are inspired by Sierpinski triangle patterns present on sea snail shells, and engineer the same kind of emergent patterns on grids of bacterial colonies. These patterns are emergent because they arise directly from one logic gate implemented in the bacteria.
Two types of bacterial colonies are placed on a grid on a millifluidic chip, in an alternating way, as shown on this picture:
Every colony is either on an ON or OFF state. When a colony of type A is ON, bacteria from this colony express GFP and produce a signaling molecule A. When a colony of type B is ON, it also expresses GFP, and produces a signaling molecule B. At the beginning of the experiment, all colonies are OFF. We induces some of the colonies of the first line, they become ON. Every colony in the second line will update its state by computing an XOR gate of the states of the two colonies above it, by sensing signals it receives from these two colonies. Once the second line has updated its state, it will send signals to the third line which will also update its state. A pattern will thus propagate line by line until the whole chip displays a pattern.
This project that combines modeling and wet-lab work will enable us to answer some questions such as how complexity can emerge from simple rules, whether it can be predicted from simple rules, how we can deal with crosstalk and leakiness of biological systems to enable a good predictability.
Second, we widen the scope of our investigation to other projects and disciplines, from scientific fields to philosophy, sociology or art. We adress once more the issue of how to deal with complexity, by interviewing experts in several fields and gathering more massive responses with a survey. We want to investigate how people deal with complexity today. Do they consider that parts are strictly ordered, and try to reduce complexity to simple parts strictly following a set of deterministic rules, or do they accept that complexity comprises a mix of order and disorder, that a part of uncertainty can't be neglected and that complex systems should be studied as a whole ? Both approaches have their advantages and their disadvantages, which one should we choose to deal with the increasing complexity of our world ?
Implementation in E. coli
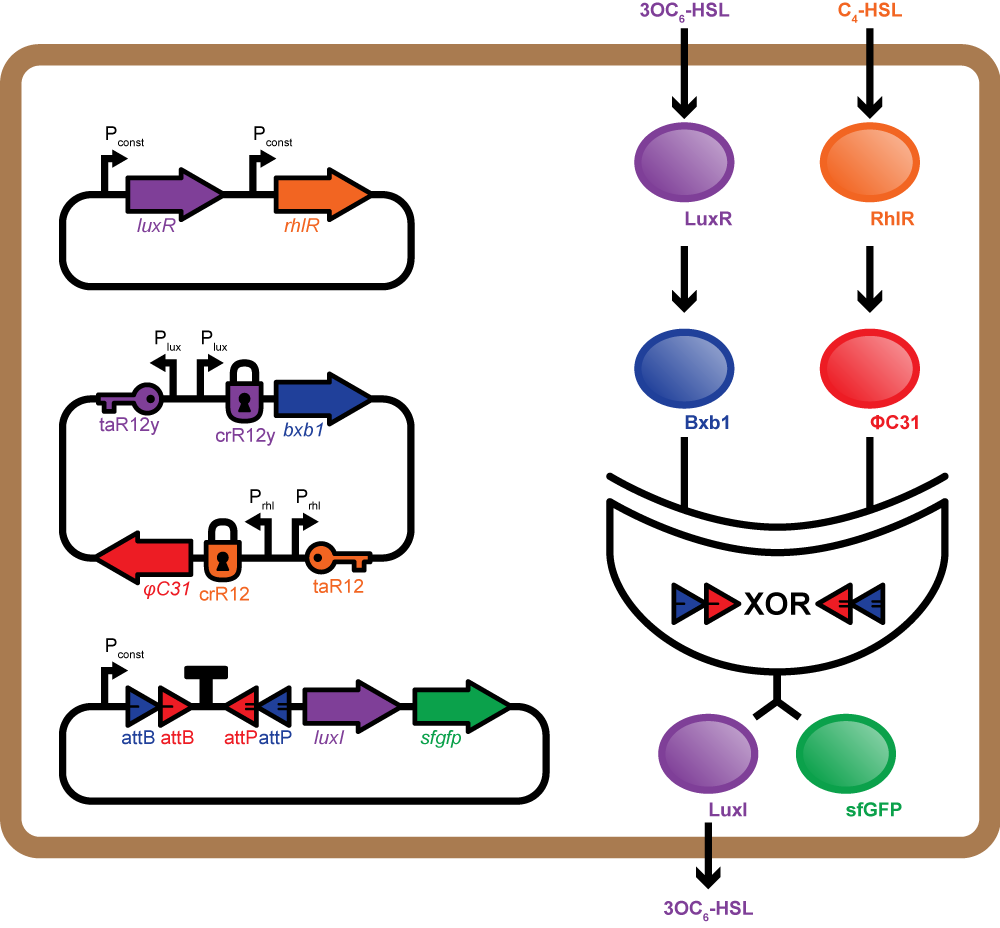
Mosaicoli involves three main constructs per cell, one for quorum sensing, one for production of integrases, and the last with the integrase based XOR logic gate to perform computation. Each cell can receive two orthogonal AHLs - the RhlI and LuxI products. The Rhl-AHL or the Lux-AHL received by the cell bind to their corresponding receptor proteins RhlR and LuxR, thus activating them. The LuxR-Lux-AHL or RhlR-Rhl-AHL complex bind to the corresponding promoters Prhl and Plux on the second plasmid and positively regulate the expression of two integrases phiC31 and Bxb1 respectively. Additionally, we use riboregulators to control the expression of the integrases thus reducing the leakiness of promoters Plux or Prhl[10].
The XOR gate present on the third plasmid comprises an asymmetric transcription terminator flanked by two pairs of opposing recombination sites recognised by phiC31 and Bxb1 respectively. gfp and luxI (or rhlI) genes are adjacent to the XOR gate and under control of the same promoter. In the absence of both integrases, the terminator blocks transcription. Expression of either integrase alone inverts the DNA encoding the terminator and allows transcription of luxI (or rhlI) and gfp. Presence of both integrases inverts the terminator twice bringing it back to its original orientation. Thus, transcription is blocked again[9].
Colonies of such cells are placed in a grid in a 3D-printed millifluidic chip. Each colony can exist in one of two states - ON and OFF. The cells are OFF if they do not produce any GFP and LuxI (or RhlI) and ON when they produce GFP and LuxI (or RhlI). The LuxI or RhlI expressed catalyse the production of the corresponding AHL molecules which diffuse out and are propagated to the colonies in the next row. Each colony updates its state by integrating signals from its neighbours (colonies in the previous rows). We expect to see complex fluorescent patterns, such as the Sierpinski triangles after several rows of colonies on the grid have updated their states.
Biological tools: quorum sensing and integrases
Quorum sensing - Bacteria, usually found in multi-species neighborhoods with billions of cells, where thought to live and divide isolated from its surroundings. However, bacteria do not try to out-compete the neighbors by mindless exponential growth but can communicate via cell-to-cell communications in order to coordinate gene-expression and as a result the overall behavior of the population. This communicative endeavor is commonly described as quorum sensing and based on the synthesis, secretion and diffusion of small molecules or 'autoinducers'.
One of the best studied quorum sensing models was found in the gram-negative bacterium Vibrio fischeri in the early 1980s[4]. Depending on the cell density and the corresponding autoinducer concentration, communication between the bacteria induces bioluminescence in the whole population. The small molecule responsible for this behavior was found to be N-(3-oxohexanoyl)homoserine lactone, which is part of the N-acetylhomoserine lactone (AHL) family. This AHL is synthesized by an enzyme called LuxI, diffuses through cell membranes and can then reach the cytosol of neighboring cells. There it binds to and activates LuxR, a protein positively regulating gene transcription. Subsequently to the investigation of V. fischeri, several other AHLs (with specific acyl side chains) and corresponding LuxR/LuxI like systems were found in various bacterial species, coordinating diverse responses including plasmid transfer, virulence, and antibiotic resistance[5].
Here we focus in particular on the quorum sensing circuitry of V. fischeri and Pseudomonas aeruginosa, another gram-negative bacteria. The additional systems are called LasR/LasI and RhlR/RhlI, also employing AHL molecules as autoinducers and are similar to the prototypical LuxR/LuxI system. Even though a key feature of quorum sensing molecules and its receptors is high specificity designed for intraspecies communication, the structural-similarity of the different AHL molecules can cause unwanted gene expression. This cross-talk has to be taken into account when re-engineering LuxR/LuxI like systems for synthetic modules and circuits[6].
Integrases -- Site specific serine recombinases, or integrases, are enzymes recognizing pairs of short and non-identical DNA sequences. Within those sites they irreversibly catalyze the excision or unidirectional inversion of DNA bases, depending on the orientation of the recognition sequence (often referred to as attB or attP). Many recombinases originate from phages integrating their genes into bacterial host genomes, hence they often do not require specific co-factors and are fully functional when heterologously expressed in bacteria. As a result, they can be employed as molecular tools in biotechnology[7].
Due to the site-specificity, recombinases like Bxb1 and PhiC31 can be employed orthogonally in synthetic systems. In addition, the combination of multiple recombinases -and their corresponding specific recognition sites- with gene regulatory sequences like promoters and terminators, allowed the engineering of Boolean logic gates[8]. For example, when an asymmetric terminator sequence is placed within a pair of recombination sites, the RNA polymerase can only proceed along this sequence if exactly one recombinase is available. Otherwise the terminator is blocking gene transription by default, or due to the repeated inversion, is again fully function as soon as both recombinases are present[9]. Such recombinase-based logic gates are at the core of our genetic circuits.
Applications and Outlook
Future development of synthetic biological systems will require the implementation of reliable synthetic circuits. These gene circuits will be designed to program new biological behaviour, dynamics and logic control.[1] A crucial part to coordinate and control the biological computation needed to perform their tasks can be seen in multichannel orthogonal communication. In order to achieve the goals of our project we need to thoroughly characterize available components, improve their performance, and develop novel constructs that get closer to actual multichannel orthogonal communication and subsequent logic processing. With our findings we will try to make a contribution to the field of biological computation by enabling further development of complex systems that rely on the interaction between its subparts.
These biological computers in turn also need to find a way to be implemented in patients, if they are developed to perform therapeutic tasks. A promising application of such synthetic biological systems is cell based therapy by alginate-microencapsulated implants.[2][3] We will try to achieve communication and input integration in a grid of alginate beads and thus producing a more complex behavior than a single bead could show. This can be seen as a step towards the development of artificial organs composed of multiple alginate-microencapsulated implants that work together to tackle tasks a single implant, which is not communicating with its neighbors, could not solve.
 "
"