Team:Aachen/Project/Gal3
From 2014.igem.org
(→Achievements) |
|||
| Line 13: | Line 13: | ||
<!-- Overview --> | <!-- Overview --> | ||
| - | <li><a href="https://2014.igem.org/Team:Aachen/Project/Gal3# | + | <li><a href="https://2014.igem.org/Team:Aachen/Project/Gal3#naturalfunctions" style="color:black"> |
<div class="team-item team-info" > | <div class="team-item team-info" > | ||
| - | + | <div class="menukachel">Natural Functions</div> | |
<!-- <br/><br/> | <!-- <br/><br/> | ||
<b>Principle of Operation</br> | <b>Principle of Operation</br> | ||
| Line 21: | Line 21: | ||
click for more information --> | click for more information --> | ||
</div> | </div> | ||
| - | <div class="team-item team-img" style="background: url(https://static.igem.org/mediawiki/2014/7/ | + | <div class="team-item team-img" style="background: url(https://static.igem.org/mediawiki/2014/7/76/Aachen_14-10-13_Galectin-3_iNB.png); norepeat scroll 0% 0% transparent; background-size:100%"> </div></a> |
</li> | </li> | ||
| - | <li><a href="https://2014.igem.org/Team:Aachen/Project/Gal3# | + | <li><a href="https://2014.igem.org/Team:Aachen/Project/Gal3#alternativesensing" style="color:black"> |
<div class="team-item team-info" > | <div class="team-item team-info" > | ||
| - | + | <div class="menukachel" style="line-height:1.5em">An Alternative Sensing Molecule</div> | |
<!-- <br/><br/> | <!-- <br/><br/> | ||
<b>Principle of Operation</br> | <b>Principle of Operation</br> | ||
| Line 32: | Line 32: | ||
click for more information --> | click for more information --> | ||
</div> | </div> | ||
| - | <div class="team-item team-img" style="background: url(https://static.igem.org/mediawiki/2014/7/ | + | <div class="team-item team-img" style="background: url(https://static.igem.org/mediawiki/2014/7/74/Aachen_14-10-13_Galectin-3-YFP_iNB.png); norepeat scroll 0% 0% transparent; background-size:100%"> </div></a> |
</li> | </li> | ||
| Line 49: | Line 49: | ||
</html> | </html> | ||
| + | |||
| + | {{Team:Aachen/BlockSeparator}} | ||
| + | |||
| + | [[File:Aachen_14-10-13_Galectin-3_iNB.png|150px|right]] | ||
| + | |||
| + | = Natural Functions of Galectin-3 = | ||
| + | <span class="anchor" id="naturalfunctions"></span> | ||
| + | |||
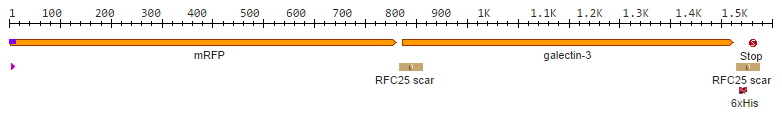
| + | Galectins are proteins of the lectin family, which possess carbohydrate recognition domains '''binding specifically to β-galactoside sugar residues'''. In humans, 10 different galantines have been identified, among which is galectin-3. | ||
| + | Galectin-3 has a size of about 31 kDA and is encoded by a single gene, LGALS3. It is found to have many physiological functions, such as '''cell adhesion, cell growth and differentiation''', and contributes to the development of '''cancer, inflammation, fibrosis''' and others. | ||
| + | |||
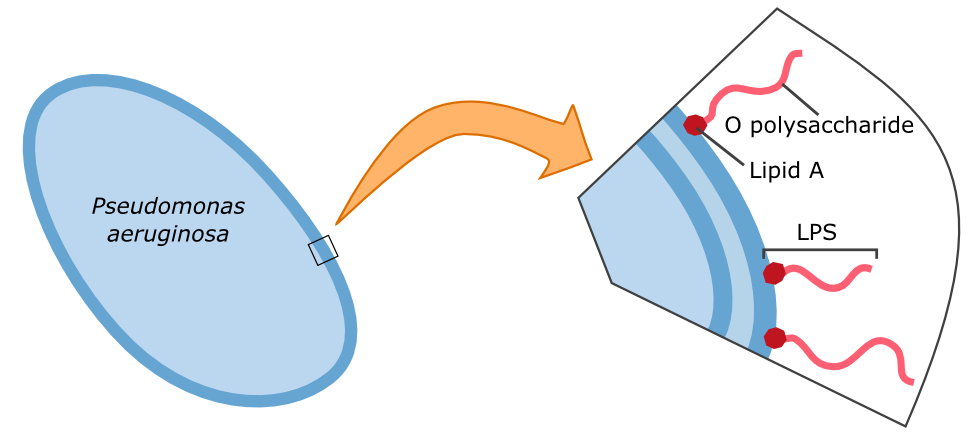
| + | Human galectin-3 is a protein of the lectin-family that was shown to bind the LPS of multiple human pathogens. Some of them, including ''P. aeruginosa'' protect themselves against the human immune system by mimicking the lipopolysaccharides (LPS) present on human erythrocytes. | ||
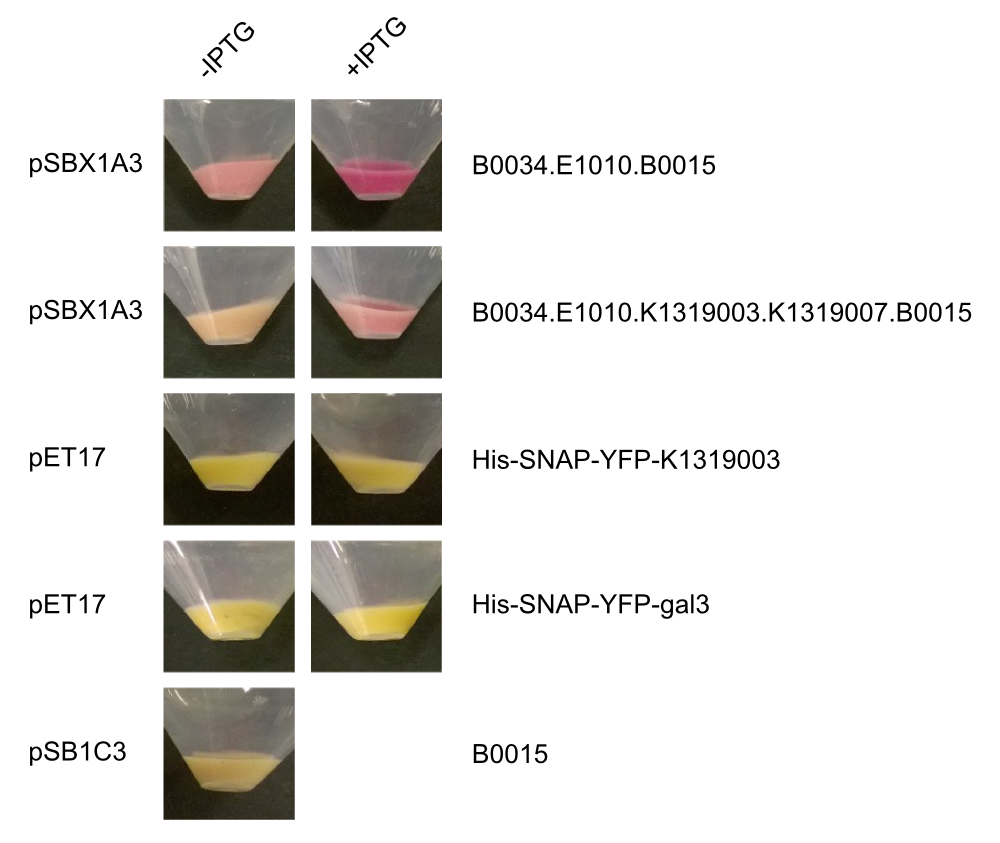
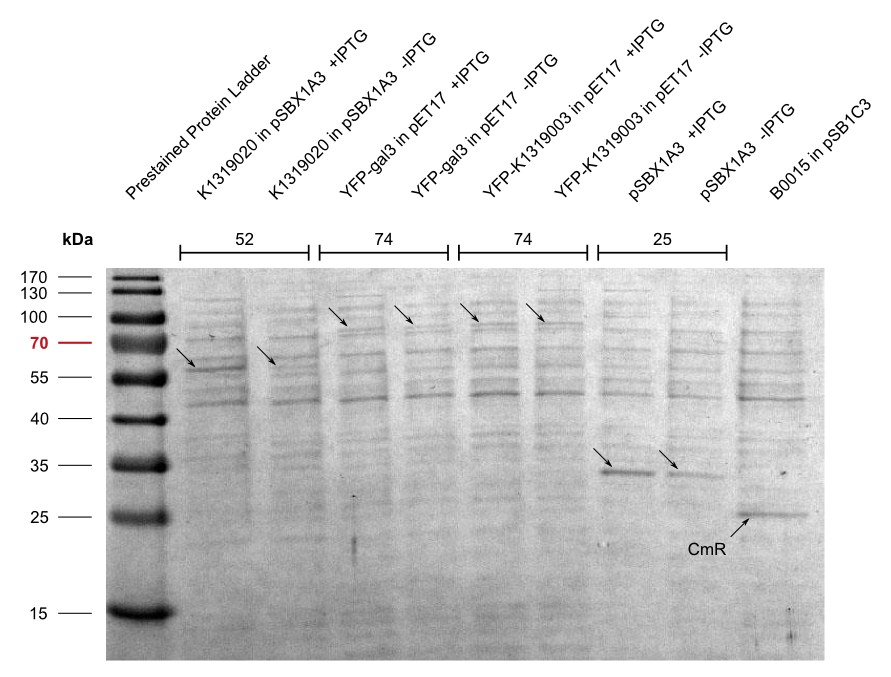
| + | By making fusion proteins of galectin-3 with fluorescent reporter proteins, pathogens can be labelled and '''made visible by fluorescence-microscopy'''. | ||
{{Team:Aachen/BlockSeparator}} | {{Team:Aachen/BlockSeparator}} | ||
| Line 64: | Line 77: | ||
{{Team:Aachen/FigureFloatRight|align=center|Aachen_14-10-09_Cell_Free_Biosensor_iNB.png|width=500px}} | {{Team:Aachen/FigureFloatRight|align=center|Aachen_14-10-09_Cell_Free_Biosensor_iNB.png|width=500px}} | ||
| - | |||
| - | |||
| - | |||
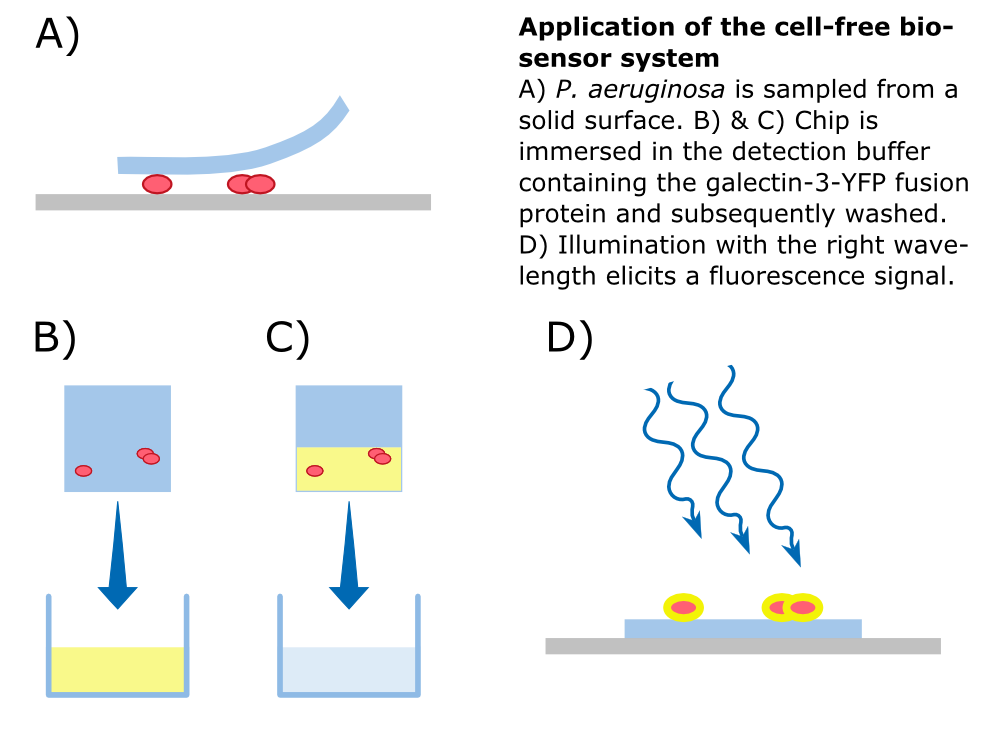
To detect ''P. aeruginosa'' cells, an agar chip could be used to sample a solid surface. However, other materials but agar can be considered to collect the pathogens. The cell stick to the sampling chip which is then immersed in a detection buffer containing the galectin-3-YFP fusion protein. Excess protein is removed during washing in a suitable buffer. The galectin-3 remains bound to the pathogen and '''illumination with 514 nm''', the excitation frequency of YFP, in a modified version of our measurement device reveals the location of the cells. The picture taken by the measurement device can then be analyzed by our software ''Measurarty''. | To detect ''P. aeruginosa'' cells, an agar chip could be used to sample a solid surface. However, other materials but agar can be considered to collect the pathogens. The cell stick to the sampling chip which is then immersed in a detection buffer containing the galectin-3-YFP fusion protein. Excess protein is removed during washing in a suitable buffer. The galectin-3 remains bound to the pathogen and '''illumination with 514 nm''', the excitation frequency of YFP, in a modified version of our measurement device reveals the location of the cells. The picture taken by the measurement device can then be analyzed by our software ''Measurarty''. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Revision as of 01:57, 18 October 2014
|
|
|
|
 "
"