Team:Aachen/Project/Model
From 2014.igem.org
AZimmermann (Talk | contribs) (→Modeling) |
AZimmermann (Talk | contribs) |
||
| Line 13: | Line 13: | ||
</center> | </center> | ||
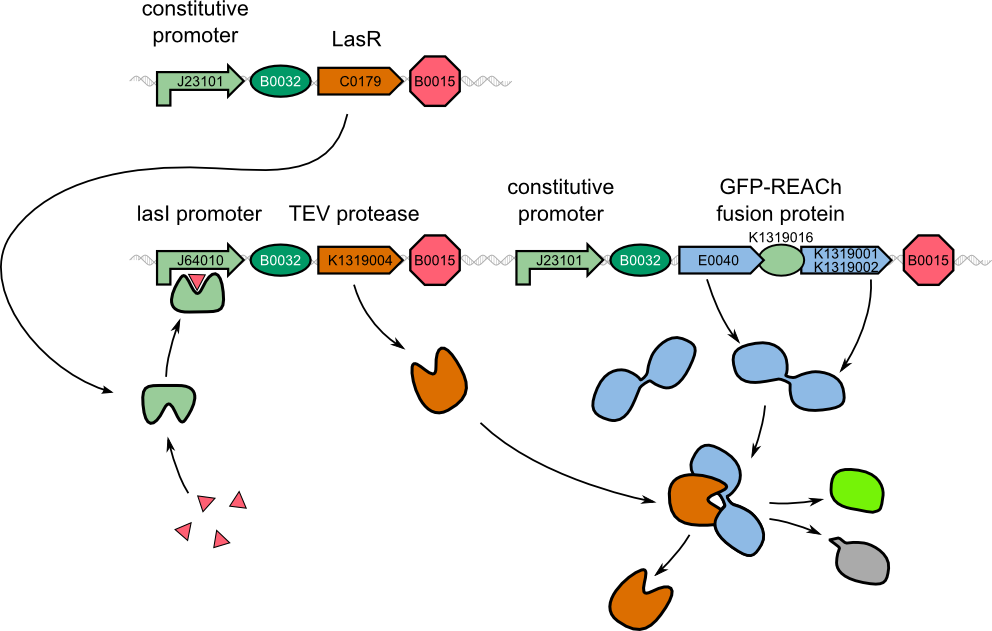
| - | For our two-dimensional biosensor, we thought of different methods to generate faster and stronger fluorescence responses from weak | + | For our two-dimensional biosensor, we thought of different methods to generate faster and stronger fluorescence responses from weak promoters. We were inspired by a recently published engineered ''dark quencher'', called REACh, that is able to extinguish the fluorescence of GFP. In our system, we wanted a fusion protein of GFP with the dark quencher to be cleaved by the very specific TEV protease that would then be introduced behind the weak quorum sensing promoter. |
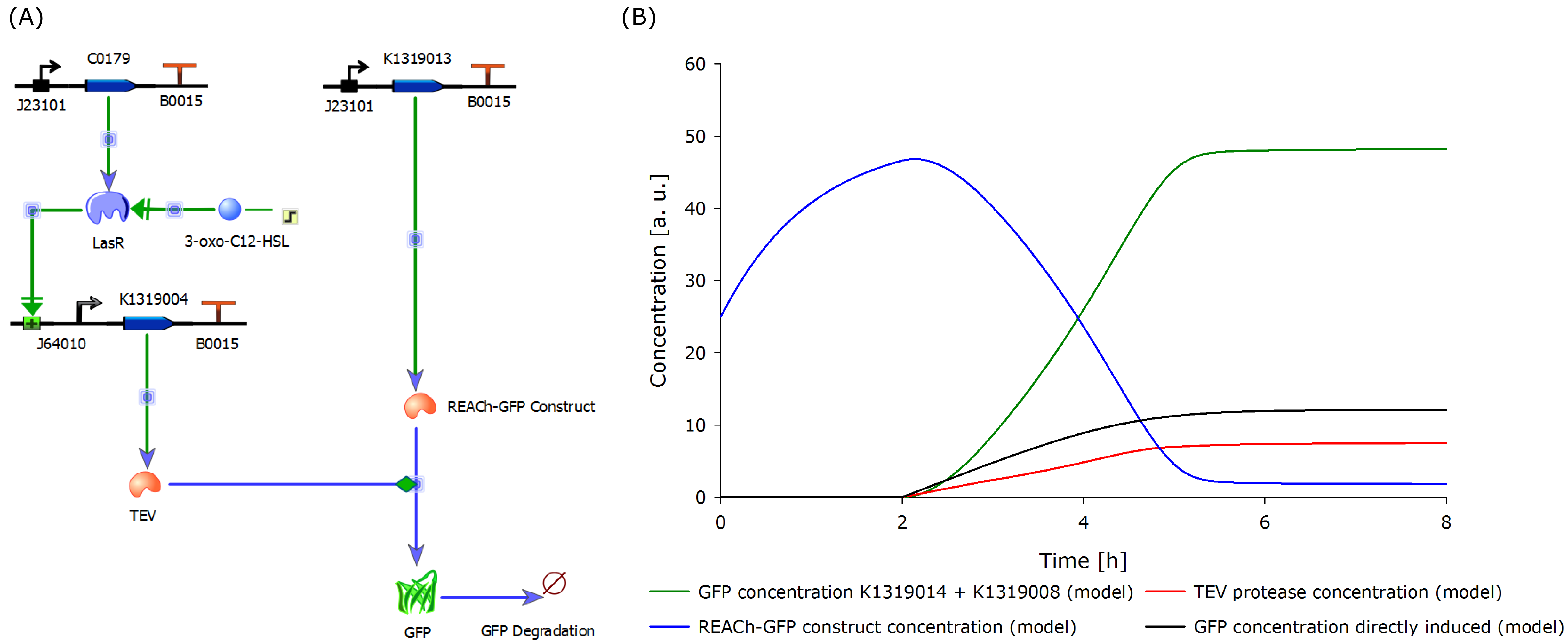
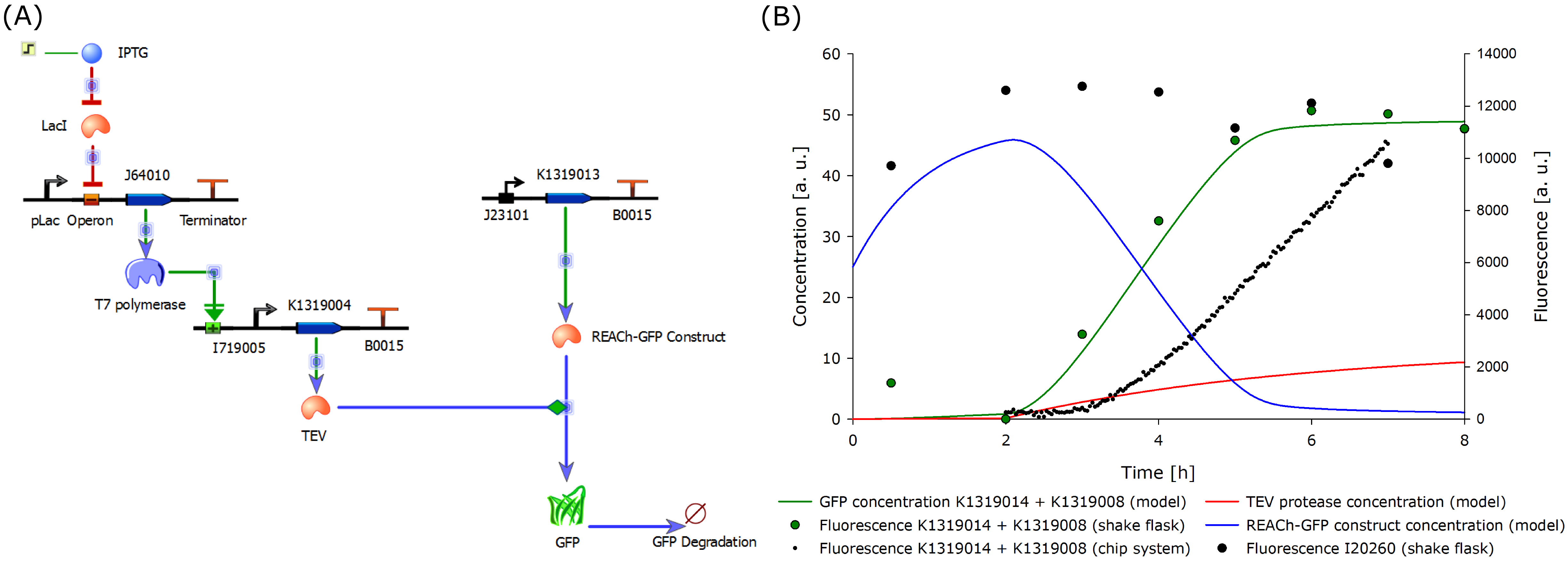
To determine if this idea was actually feasible, we decided to model the system using the CAD tool TinkerCell (Chandran, Bergmann and Sauro, 2009). | To determine if this idea was actually feasible, we decided to model the system using the CAD tool TinkerCell (Chandran, Bergmann and Sauro, 2009). | ||
| - | To compare the response time of the fluorescence signal between our theoretical system and a traditional biosensor, we included a direct expression of GFP in the same plot (below). In the results shown below, the strength of the | + | To compare the response time of the fluorescence signal between our theoretical system and a traditional biosensor, we included a direct expression of GFP in the same plot (below). In the results shown below, the strength of the promoter used for the direct GFP expression (traditional approach) is even twice as high as the strength of the promoter upstream of the TEV coding sequence in our new approach. Despite the weaker promoter, a '''higher GFP concentration is generated in the model of the novel biosensor''', predicting a quicker response time of our system. |
Revision as of 23:25, 17 October 2014
|
|
 "
"