Team:Aachen/Project/FRET Reporter
From 2014.igem.org
AZimmermann (Talk | contribs) (→Determining the Quenching ability of REACh1 and REACh2) |
AZimmermann (Talk | contribs) |
||
| Line 88: | Line 88: | ||
<span class="anchor" id="fluorescence"></span> | <span class="anchor" id="fluorescence"></span> | ||
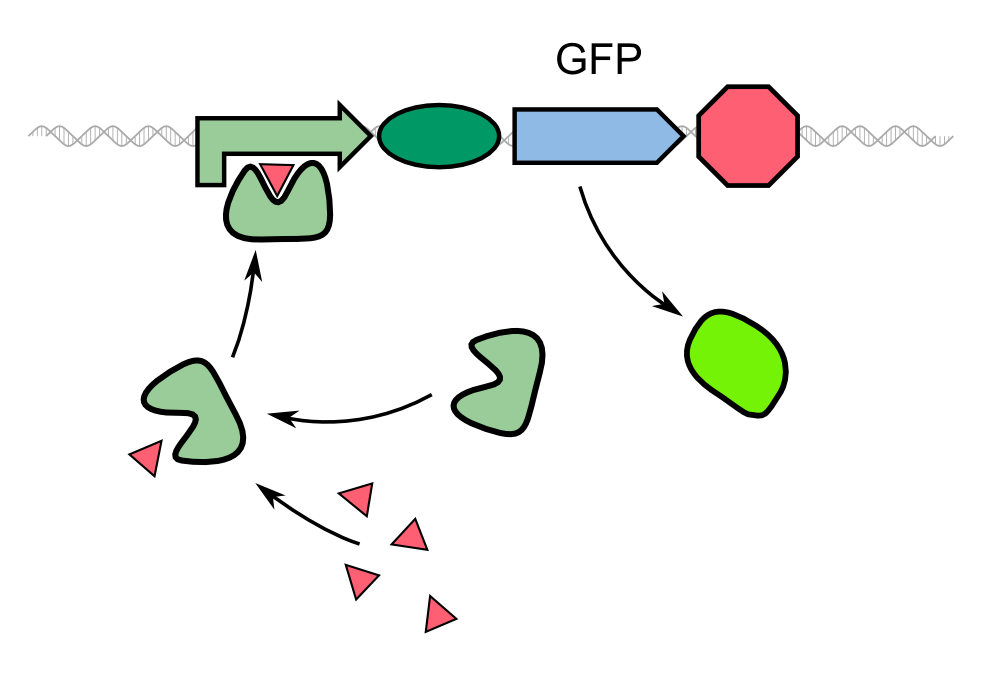
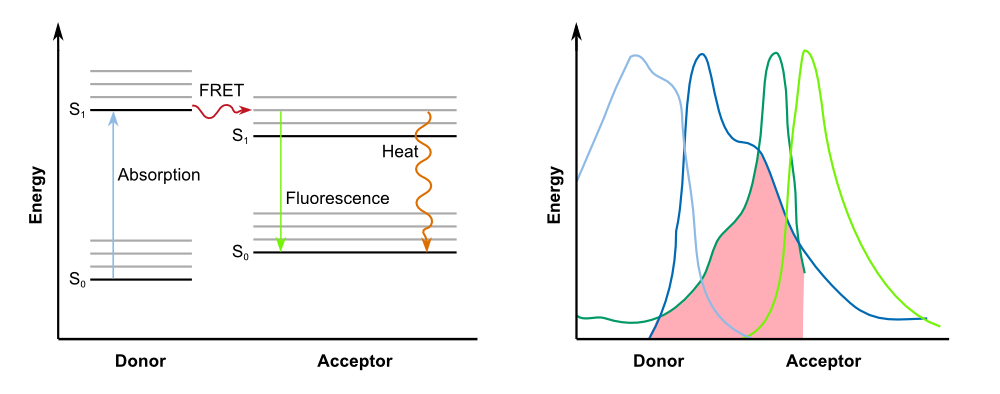
| - | Biosensors often work with a system that comprises a reported gene under the control of a | + | Biosensors often work with a system that comprises a reported gene under the control of a promoter directly induced by the chemical that the sensor is supposed to detect. In the case of our 2D biosensor for ''Pseudomonas aeruginosa'', the expression of our reporter gene, GFP, would be directly induced by the activity of the bacterium's quorum sensing molecules. However, transcription, translation, folding and post-translational modifications take their time. Since our goal is to detect the pathogen as fast as possible, we wanted to use a system that gives a fluorescent answer faster than just expressing the fluorescent protein. |
<center> | <center> | ||
| - | {{Team:Aachen/Figure|Aachen_Traditional_biosensor.png|align=center|title=Schematic model of a traditional biosensor|subtitle=In this model, the expression of GFP is directly controlled by a | + | {{Team:Aachen/Figure|Aachen_Traditional_biosensor.png|align=center|title=Schematic model of a traditional biosensor|subtitle=In this model, the expression of GFP is directly controlled by a promoter whose activator binds to a molecule secreted by the pathogen.|width=500px}} |
</center> | </center> | ||
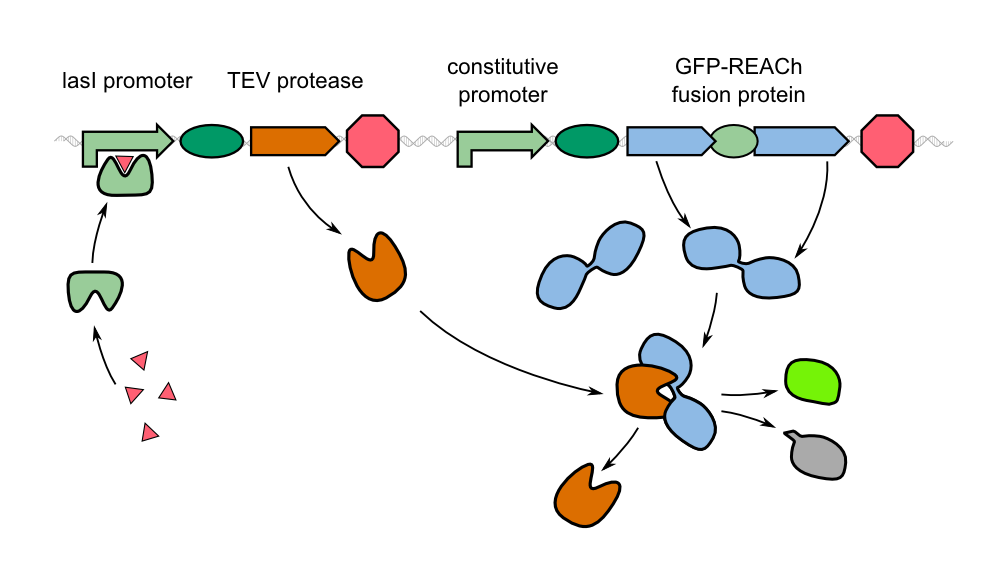
| - | As an alternative to the traditional approach, we '''constitutively express our reporter gene in a quenched form'''. in the GFP-REACh fusion protein fluorescence is suppressed. Our biosensor gives a response when homoserine lactones of ''P. aeruginosa'' are taken up by our sensor cells where the '''autoinducer activates the expression of the TEV protease''' by binding to the LasI | + | As an alternative to the traditional approach, we '''constitutively express our reporter gene in a quenched form'''. in the GFP-REACh fusion protein fluorescence is suppressed. Our biosensor gives a response when homoserine lactones of ''P. aeruginosa'' are taken up by our sensor cells where the '''autoinducer activates the expression of the TEV protease''' by binding to the LasI promoter in front of the protease gene. |
<center> | <center> | ||
| Line 148: | Line 148: | ||
<span class="anchor" id="gfp-reach"></span> | <span class="anchor" id="gfp-reach"></span> | ||
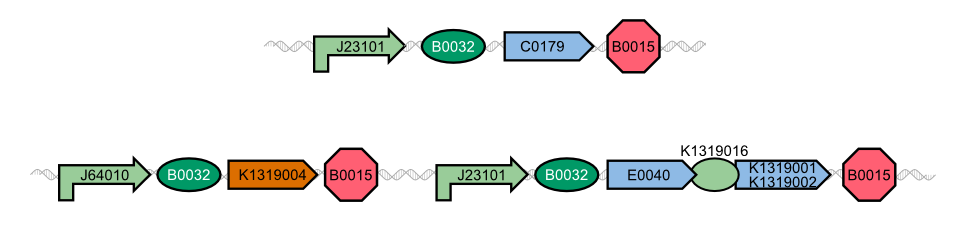
| - | In our project, we reproduced the REACh1 and REACh2 proteins by subjecting an RFC-25 compatible version of the BioBrick [http://parts.igem.org/Part:BBa_E0030 E0030] (EYFP) to a '''QuikChange mutation''', creating the BioBricks [http://parts.igem.org/Part:BBa_K1319001 K1319001] and [http://parts.igem.org/Part:BBa_K1319002 K1319002]. Subsequently, we fused each REACh protein with '''GFP (mut3b)''' which is available as BioBrick [http://parts.igem.org/Part:BBa_E0040 E0040]. The protein complex was linked via a '''protease cleavage site''', [http://parts.igem.org/Part:BBa_K1319016 K1319016]. As constitutive | + | In our project, we reproduced the REACh1 and REACh2 proteins by subjecting an RFC-25 compatible version of the BioBrick [http://parts.igem.org/Part:BBa_E0030 E0030] (EYFP) to a '''QuikChange mutation''', creating the BioBricks [http://parts.igem.org/Part:BBa_K1319001 K1319001] and [http://parts.igem.org/Part:BBa_K1319002 K1319002]. Subsequently, we fused each REACh protein with '''GFP (mut3b)''' which is available as BioBrick [http://parts.igem.org/Part:BBa_E0040 E0040]. The protein complex was linked via a '''protease cleavage site''', [http://parts.igem.org/Part:BBa_K1319016 K1319016]. As constitutive promoter we use [http://parts.igem.org/Part:BBa_J23101 J232101]. When GFP is connected to either REACh quencher, GFP will absorb light but the energy will be transferred to REACh via FRET and is then emitted in the form of heat; the fluorescence is quenched. Our cells also constitutively express the '''[http://parts.igem.org/Part:BBa_C0179 LasR] activator'''. Together with the HSL molecules from ''P. aeruginosa'', LasR binds to the '''[http://parts.igem.org/Part:BBa_J64010 LasI] promoter''' that controls the expression of the TEV protease, which we make available as [http://parts.igem.org/Part:BBa_K1319004 K1319004]. When the fusion protein is cleaved by the TEV protease, REACh will be separated from GFP. The latter will then be able to absorb and emit light as usual. |
{{Team:Aachen/Figure|align=center|Aachen_14-10-08_REACh_approach_with_BioBricks_iNB.png|title= Composition of our biosensor|subtitle=For our biosensor, we use a mix of already available and self-constructed BioBricks.|width=900px}} | {{Team:Aachen/Figure|align=center|Aachen_14-10-08_REACh_approach_with_BioBricks_iNB.png|title= Composition of our biosensor|subtitle=For our biosensor, we use a mix of already available and self-constructed BioBricks.|width=900px}} | ||
| Line 157: | Line 157: | ||
| - | {{Team:Aachen/Figure|align=center|Aachen 14-10-17 IPTG REACh iFG.png|title= Composition of our biosensor for IPTG|subtitle=To test the funtionality of our sensor concept, we expressed the TEV protease with a T7 | + | {{Team:Aachen/Figure|align=center|Aachen 14-10-17 IPTG REACh iFG.png|title= Composition of our biosensor for IPTG|subtitle=To test the funtionality of our sensor concept, we expressed the TEV protease with a T7 promoter.|width=900px}} |
{{Team:Aachen/BlockSeparator}} | {{Team:Aachen/BlockSeparator}} | ||
| Line 184: | Line 184: | ||
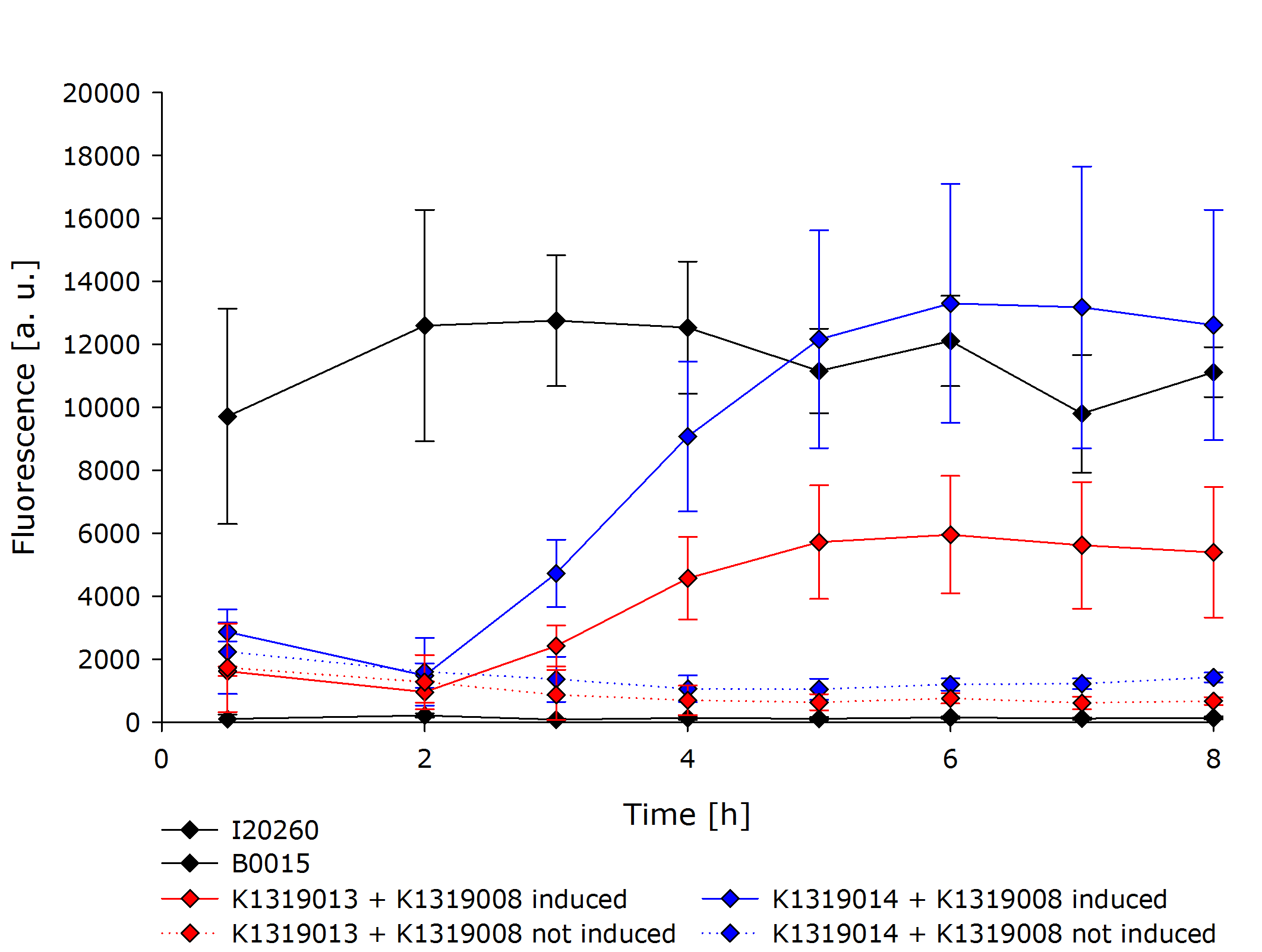
The characterization of the TEV protease and the REACh1 and REACh2 dark quenchers was performed by introducing both simultaneously into ''E. coli'' Bl21 (DE3). The resulting double plasmid cells therefore contained [http://parts.igem.org/Part:BBa_K1319013 K1319013] (GFP-REACh1 fusion protein) or [http://parts.igem.org/Part:BBa_K1319014 K1319014] (GFP-REACh2 fusion protein) and [http://parts.igem.org/Part:BBa_K1319008 K1319008] (IPTG-inducible TEV protease). K1319013 and K1319014 were located on a pSB3K3 plasmid backbone and K1319008 on a pSB1C3 backbone, two standard iGEM plasmids with different oris allowing simultaneous use in one cell. | The characterization of the TEV protease and the REACh1 and REACh2 dark quenchers was performed by introducing both simultaneously into ''E. coli'' Bl21 (DE3). The resulting double plasmid cells therefore contained [http://parts.igem.org/Part:BBa_K1319013 K1319013] (GFP-REACh1 fusion protein) or [http://parts.igem.org/Part:BBa_K1319014 K1319014] (GFP-REACh2 fusion protein) and [http://parts.igem.org/Part:BBa_K1319008 K1319008] (IPTG-inducible TEV protease). K1319013 and K1319014 were located on a pSB3K3 plasmid backbone and K1319008 on a pSB1C3 backbone, two standard iGEM plasmids with different oris allowing simultaneous use in one cell. | ||
| - | [http://parts.igem.org/Part:BBa_I20260 I20260] was used as a positive control because I20260 contains the same | + | [http://parts.igem.org/Part:BBa_I20260 I20260] was used as a positive control because I20260 contains the same promoter ([http://parts.igem.org/Part:BBa_J23101 J23101]), the same RBS ([http://parts.igem.org/Part:BBa_B0032 B0032]) and the same version of GFP ([http://parts.igem.org/Part:BBa_E0040 E0040]) and is located on the same plasmid backbone pSB3K3. Therefore, it is expected that when all fusion proteins are successfully cut by the TEV protease, the fluorescence level of the double plasmid constructs reaches the same level as the positive control of I20260. As a negative control [http://parts.igem.org/Part:BBa_B0015 B0015] was used, a coding sequence of a terminator which should not show any sign of fluorescence. |
To characterize our REACh1/2 constructs in combination with the TEV protease a growth experiment was conducted. Both of the double plasmid constructs, a constitutive expression of GFP (I20260) as positive control and B0015 as negative control were compared. For each expression IPTG-induced and non-induced cultures were grown in parallel. All measurements were done in a biological triplicate. | To characterize our REACh1/2 constructs in combination with the TEV protease a growth experiment was conducted. Both of the double plasmid constructs, a constitutive expression of GFP (I20260) as positive control and B0015 as negative control were compared. For each expression IPTG-induced and non-induced cultures were grown in parallel. All measurements were done in a biological triplicate. | ||
| Line 194: | Line 194: | ||
The negative control B0015 did not exhibit any significant fluorescence. The positive control I20260 showed a steady level of fluorescence as expected due to the constitutive expression of GFP. As expected, the production is also independent of addition of IPTG therefore the triplicates have been merged together. | The negative control B0015 did not exhibit any significant fluorescence. The positive control I20260 showed a steady level of fluorescence as expected due to the constitutive expression of GFP. As expected, the production is also independent of addition of IPTG therefore the triplicates have been merged together. | ||
| - | Both double plasmid constructs K1319013 + K1319008 and K1319014 + K1319008 did not exhibit a strong fluorescence before induction with IPTG. In the non-induced state, the fluorescence stays low and does not increase over time. It is significantly weaker than the fluorescence reached by the induced constructs or the positive control but was higher than the negative control. The higher base level of fluorescence in the not induced constructs is due to the imperfect quenching and the leakiness of the IPTG inducible | + | Both double plasmid constructs K1319013 + K1319008 and K1319014 + K1319008 did not exhibit a strong fluorescence before induction with IPTG. In the non-induced state, the fluorescence stays low and does not increase over time. It is significantly weaker than the fluorescence reached by the induced constructs or the positive control but was higher than the negative control. The higher base level of fluorescence in the not induced constructs is due to the imperfect quenching and the leakiness of the IPTG inducible promoter. |
The induced double plasmid constructs exhibited a fast rise in fluorescence after induction. '''The signal strenght increased ~ 10-fold over the non-induced constructs.''' K1319014 + K1319008 reached the the same level of fluorescence as I20260, indicating a complete cleavage of the fusion proteins by the TEV protease. K1319013 + K1319008 did not reach a level of fluorescence as high as K1319013 + K1319008, however, the nearly 10-fold increase in fluorescence after induction is a clear indicator for the TEV protease cutting the fusion protein K1319013. The weaker fluorescence signal is probably due to a lower expression level of K1319013 in the cells compared to the expression level of K1319014. | The induced double plasmid constructs exhibited a fast rise in fluorescence after induction. '''The signal strenght increased ~ 10-fold over the non-induced constructs.''' K1319014 + K1319008 reached the the same level of fluorescence as I20260, indicating a complete cleavage of the fusion proteins by the TEV protease. K1319013 + K1319008 did not reach a level of fluorescence as high as K1319013 + K1319008, however, the nearly 10-fold increase in fluorescence after induction is a clear indicator for the TEV protease cutting the fusion protein K1319013. The weaker fluorescence signal is probably due to a lower expression level of K1319013 in the cells compared to the expression level of K1319014. | ||
| Line 207: | Line 207: | ||
===Determining the Quenching ability of REACh1 and REACh2=== | ===Determining the Quenching ability of REACh1 and REACh2=== | ||
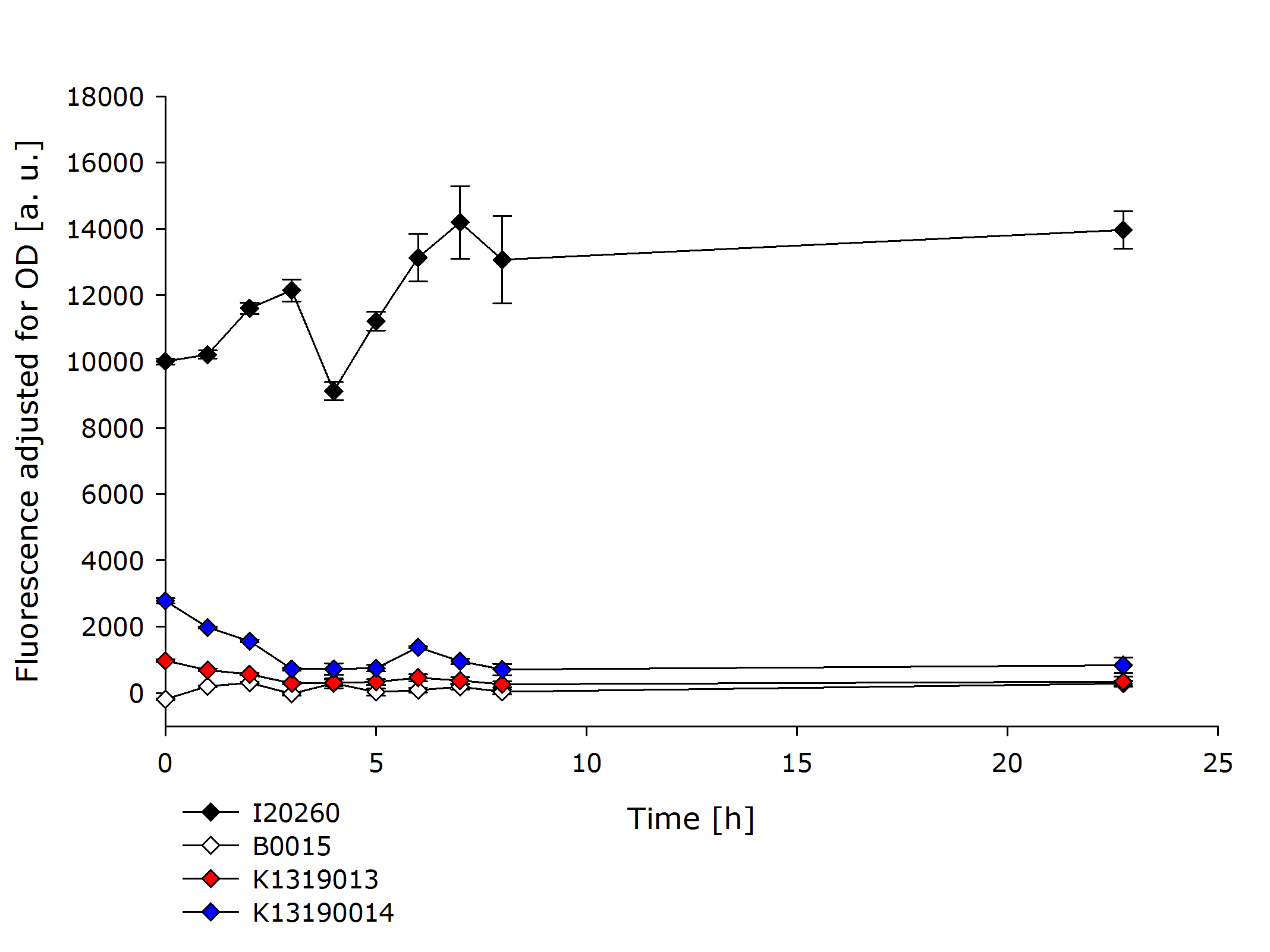
| - | In order to further evaluate the quenching ability of the REACh1 and REACh2 constructs in the fusion proteins produced by K1319013 and K1319014, they were expressed alone without an IPTG inducible TEV protease. This eliminated the effect of a potential leakiness of the non induced | + | In order to further evaluate the quenching ability of the REACh1 and REACh2 constructs in the fusion proteins produced by K1319013 and K1319014, they were expressed alone without an IPTG inducible TEV protease. This eliminated the effect of a potential leakiness of the non induced promoter to reliably assess the quenching ability of the REACh1 and REACh2 proteins. |
{{Team:Aachen/Figure|Aachen_K1319001_and_K1319002.PNG|title=Comparison of K1319013 and K1319014 with I20260 and B0015|subtitle=K1319013 and K1319014 show a severely reduced fluorescence compared to the positive control I20260.|width=900px}} | {{Team:Aachen/Figure|Aachen_K1319001_and_K1319002.PNG|title=Comparison of K1319013 and K1319014 with I20260 and B0015|subtitle=K1319013 and K1319014 show a severely reduced fluorescence compared to the positive control I20260.|width=900px}} | ||
| Line 221: | Line 221: | ||
===Comparing fluorescence kinetics of the GFP-REACh fusion proteins with a Standard lacI-inducible GFP Expression=== | ===Comparing fluorescence kinetics of the GFP-REACh fusion proteins with a Standard lacI-inducible GFP Expression=== | ||
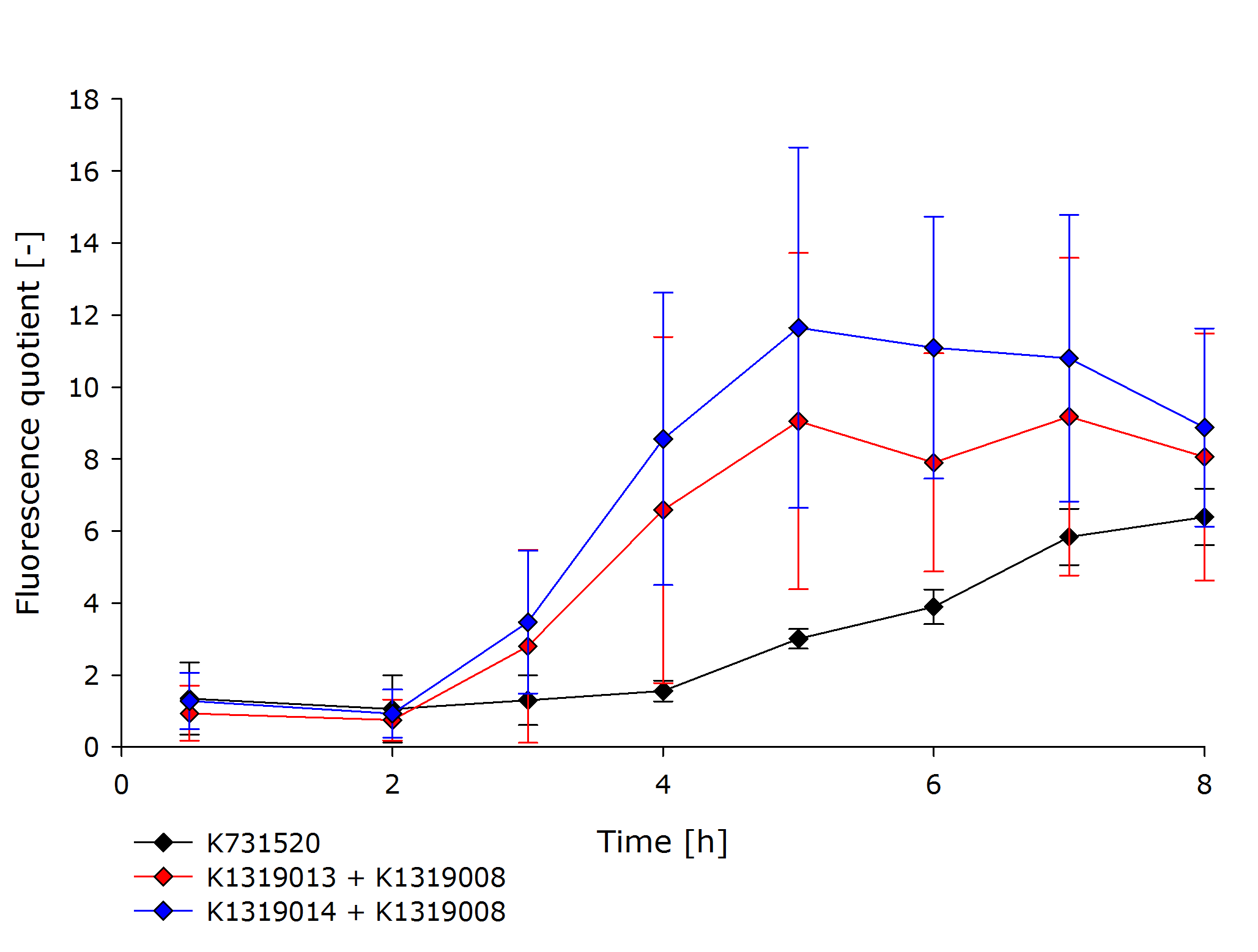
| - | To assess the kinetics of the fusion proteins K1319013 (GFP-REACh1) and K1319014 (GFP-REACh2), the double plasmid systems K1319013 + K1319008 and K1319014 + K1319008 were compared to a standard expression of GFP under the control of a lacI | + | To assess the kinetics of the fusion proteins K1319013 (GFP-REACh1) and K1319014 (GFP-REACh2), the double plasmid systems K1319013 + K1319008 and K1319014 + K1319008 were compared to a standard expression of GFP under the control of a lacI promoter in [http://parts.igem.org/Part:BBa_K731520 K731520], a BioBrick made by the iGEM Team TRENTO in 2012. This tested the hypothesis of achieving a faster fluorescence response with the GFP-REACh fusion proteins compared to a standard expression. |
K731520 and the double plasmid constructs K1319013 + K1319008 and K1319014 + K1319008 were cultivated in ''E. coli'' BL21(DE3), and fluorescence and OD was measured.The fluorescence was adjusted for the OD to show a relative fluorescence on a per cell basis. The difference between the induced and non-induced state, the fluorescence quotient, serves as a better indicator for a system used as a sensor because the difference between an ''on'' and ''off'' state is more important for a clear and unmistakable signal than the overall fluorescence. Hence, the OD-adjusted fluorescence quotient for both double plasmid constructs and K731520 was obtained and plotted in the following graph. | K731520 and the double plasmid constructs K1319013 + K1319008 and K1319014 + K1319008 were cultivated in ''E. coli'' BL21(DE3), and fluorescence and OD was measured.The fluorescence was adjusted for the OD to show a relative fluorescence on a per cell basis. The difference between the induced and non-induced state, the fluorescence quotient, serves as a better indicator for a system used as a sensor because the difference between an ''on'' and ''off'' state is more important for a clear and unmistakable signal than the overall fluorescence. Hence, the OD-adjusted fluorescence quotient for both double plasmid constructs and K731520 was obtained and plotted in the following graph. | ||
| Line 273: | Line 273: | ||
Subsequently the combined construct will be characterizes the same way as the double plasmid system and other options regarding different fluorescence proteins with different quenchers will be considered to be able to have multiple fluorescence responses readable at the same time while still being faster than normal expression. | Subsequently the combined construct will be characterizes the same way as the double plasmid system and other options regarding different fluorescence proteins with different quenchers will be considered to be able to have multiple fluorescence responses readable at the same time while still being faster than normal expression. | ||
| - | Also finding and testing different | + | Also finding and testing different promoters which can be used to express the TEV protease is planned to be able to detect not only ''Pseudomonas aeruginosa'' but also other pathogens. Also the technology can be expanded to other molecules enabling other research areas to profit from the faster fluorescence response. |
Revision as of 23:24, 17 October 2014
|
|
|
|
|
|
|
|
 "
"