Team:Aachen/Notebook/Software/Measurarty
From 2014.igem.org
(→Source Code) |
(→Achievements) |
||
| Line 246: | Line 246: | ||
<span class="anchor" id="measurartyachievements"></span> | <span class="anchor" id="measurartyachievements"></span> | ||
| - | + | Measurarty is the image analysis logic behind our project. | |
| - | + | It uses easy-to-understand construct put together into a pipeline, that is still easy to understand, easily maintainable - and if needed - easily adaptable. | |
| - | + | For example, changing from green to red fluorescence, only means to change the ''createMask'' function to select another target area. | |
| - | * | + | |
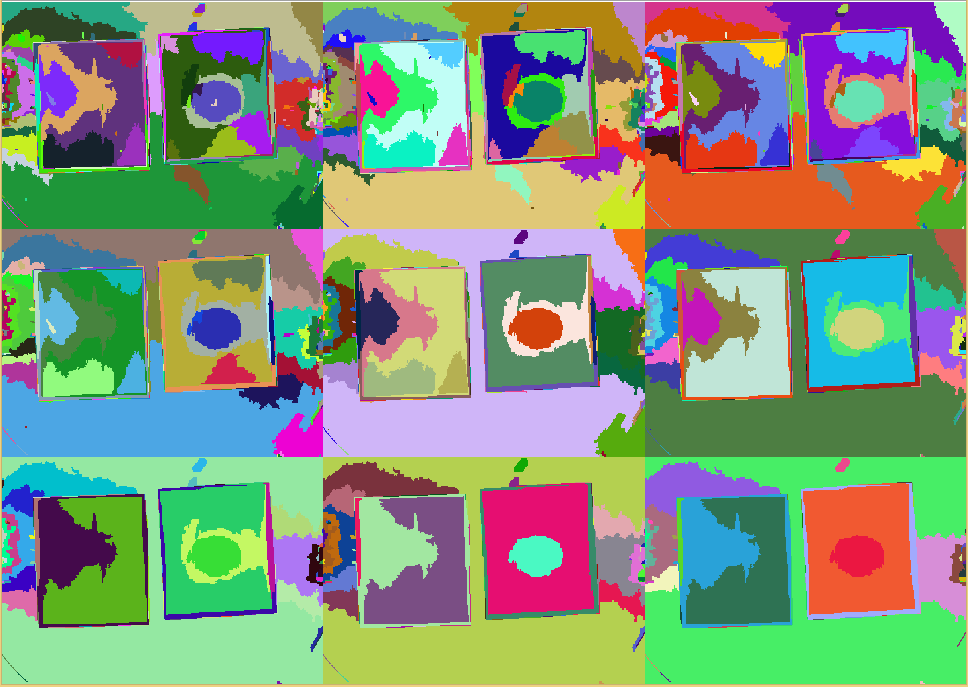
| + | Overall the results look convincing. We have not yet performed a comparison to a manual delineation, however by eye it can be seen that the results look promising to have a low error. | ||
| + | |||
| + | Talking about computational complexity, the matlab code of course performs better than our own C++ implementation, which must be regarded as a proof-of-principle. | ||
| + | |||
| + | Space-wise the code depends heavily on the image size $O( x \cdot y)$ (width $x$, height $y$, which also limits the number of edges in SRM between regions (as each pixel is one region to start with). Obviously it can not take less memory, as the image is stored in an uncompressed format. | ||
| + | |||
| + | Computational-wise our code is more interesting. The thresholding, image conversion and gradient steps are linear in the number of pixels, and thus in $O(x \cdot y)$. | ||
| + | Unfortunately the summation of the gradient for the smoothness index adds a heavy factor to it (k-neighbourhood for smoothness index). | ||
| + | Due to the merging step in our C++-SRM algorithm implementation our code unfortunately has to do $O(x^2 \cdot y^2)$ comparisons, which then finally results in a runtime complexity of $O( x^2 \cdot y^2)$. | ||
| + | |||
| + | *include image here | ||
| + | |||
| + | From the above figure it can also be seen that the detected amount of pathogenic-area correlates with time after induction. | ||
| + | The lag-phase can be explained first by the lag-phase of the cells, which first need to generate a response to the pathogene, and on the other hand by too low fluorescence which is not detectable. | ||
| + | The pixel-count also meets the expectation when looking at the sample files by eye. | ||
| + | |||
| + | It can be concluded that the measuararty pipeline defines robustly working chip-analysis algorithm which can detect pathogens from images supplied from our own device. | ||
| + | Therefore this algorithm closes the gap between our biology, detection hardware and the user, who wants easy-to-interpret results. | ||
{{Team:Aachen/BlockSeparator}} | {{Team:Aachen/BlockSeparator}} | ||
Revision as of 10:30, 17 October 2014
|
|
|
|
|
|
|
 "
"