Team:Aachen/Notebook/Software/Measurarty
From 2014.igem.org
(→Statistical Region Merging (SRM)) |
(→Measurarty) |
||
| Line 12: | Line 12: | ||
= ''Measurarty'' = | = ''Measurarty'' = | ||
| - | ''Measurarty'' | + | ''Measurarty'' is the evil player in the game of ''Cellock Holmes'' and ''WatsOn''. |
| - | Measurarty is the | + | Measurarty is the pathogen detection logic behind our project. |
| - | Using our '' | + | Using our ''Measurarty'' algorithm, we want to automatically detect pathogens from the chip photos delivered by WatsOn, without human interaction. |
Besides reducing the risk of human errors, this makes our device usable by almost everyone. | Besides reducing the risk of human errors, this makes our device usable by almost everyone. | ||
| Line 73: | Line 73: | ||
<span class="anchor" id="intro"></span> | <span class="anchor" id="intro"></span> | ||
| - | Our device control software is able to take images of incubated chips | + | Our [https://2014.igem.org/Team:Aachen/Notebook/Engineering/WatsOn#watsonsoftware device control software] is able to take images of incubated chips inside WatsOn. Yet, that does not bring the user closer to the answer of the question: |
| - | <center>''''' | + | <center>'''''What's on the chip?'''''</center> |
| - | In fact, answering this question seems trivial for a human | + | In fact, answering this question seems trivial for a human: Just check whether a colony grown has grown on the chip and you're done. This task is even easier with our chip system, because these show fluorescence wherever a pathogen has been detected. |
| - | Just check whether | + | |
| - | + | ||
| - | But is this an easy task for a computer? Actually not. The task of automatic detection is tried | + | But is this an easy task for a computer? Actually not. The task of automatic detection is tried by several disciplines in computer science, from pattern recognition over machine learning to by medical imaging chairs. |
| - | + | Here, we would like to present a pipeline for this task that makes use of '''easy segmentation and classification algorithms'''. | |
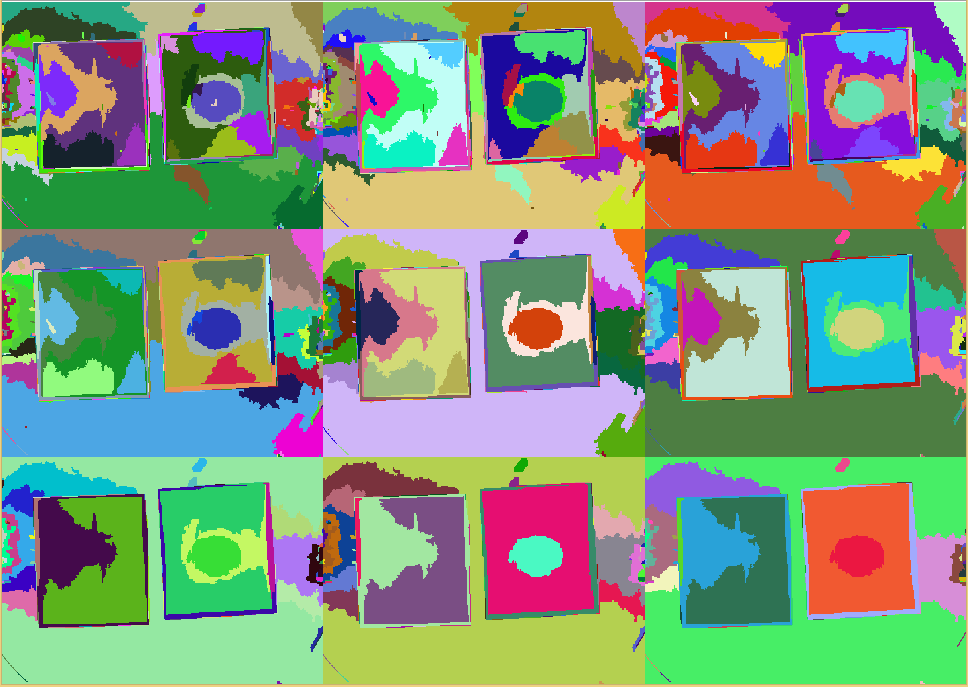
| - | First | + | First, ''Measurarty'' segments the target image using '''Statistical Region Merging (SRM)''' in order to find regions of similar properties. After this step, we can segment the picture using '''histogram thresholding''' in [http://en.wikipedia.org/wiki/HSL_and_HSV HSV color space] to find candidate regions for pathogens. |
| - | Finally a classification algorithm can detect the | + | Finally, a classification algorithm can detect the pathogen on our chips. |
{{Team:Aachen/BlockSeparator}} | {{Team:Aachen/BlockSeparator}} | ||
| Line 94: | Line 92: | ||
<span class="anchor" id="SRM"></span> | <span class="anchor" id="SRM"></span> | ||
| - | Before | + | Before briefly introducing Statistical Region Merging (SRM), we would like to explain why we need this step, and why this algorithm is an ideal choice. |
| - | Compared to other clustering algorithms, SRM is quite | + | Compared to other clustering algorithms, SRM is quite leight weight, yet delivers ''deterministic'' results and is not dependent on a certain seed (like ''k''-means, for example). |
| - | On the other hand it can create as many refinements as one wants and | + | On the other hand, it can create as many refinements as one wants and is thus flexible enough for the our purposes. Finally there's already been knowledge about this algorithm in the group. |
| - | Statistical Region Merging (SRM) | + | Statistical Region Merging (SRM) (Nook and Nielson, 2004) is a clustering technique also used directly for image segmentation. |
| - | A region $R$ is a set of pixels and the cardinality $\lvert R \rvert$ determines how many pixels are in | + | A region $R$ is a set of pixels and the cardinality $\lvert R \rvert$ determines how many pixels are in one region. |
Starting with a sorted set of connected regions (w. r. t. some distance function $f$), two regions $R$ and $R'$ are merged if the qualification criteria $\vert \overline{R'}-\overline{R} \vert \leq \sqrt{b^2(R)+b^2(R')}$ with $b(R) = g \cdot \sqrt{\frac{\ln \frac{\mathcal{R}_{\lvert R \rvert}}{\delta}}{2Q\lvert R \rvert}}$ is fulfilled. | Starting with a sorted set of connected regions (w. r. t. some distance function $f$), two regions $R$ and $R'$ are merged if the qualification criteria $\vert \overline{R'}-\overline{R} \vert \leq \sqrt{b^2(R)+b^2(R')}$ with $b(R) = g \cdot \sqrt{\frac{\ln \frac{\mathcal{R}_{\lvert R \rvert}}{\delta}}{2Q\lvert R \rvert}}$ is fulfilled. | ||
Therefore, $\mathcal{R}_{\lvert R \rvert}$ is the set of regions with $\lvert R \rvert$ pixels. | Therefore, $\mathcal{R}_{\lvert R \rvert}$ is the set of regions with $\lvert R \rvert$ pixels. | ||
| Line 141: | Line 139: | ||
<span class="anchor" id="segment"></span> | <span class="anchor" id="segment"></span> | ||
| - | In the segmentation stage all background regions get removed. This task is quite crucial. If one removes too few, the final stage of finding | + | In the segmentation stage all background regions get removed. This task is quite crucial. If one removes too few, the final stage of finding pathogens might get irritated. |
On the other hand, if one removes too many regions, positive hits might get removed early before detection. This surely also must be avoided. | On the other hand, if one removes too many regions, positive hits might get removed early before detection. This surely also must be avoided. | ||
| Line 225: | Line 223: | ||
\end{equation} | \end{equation} | ||
| - | Using a thresholding, $TS_l \leq s(p) \leq TS_u \wedge TI_l \leq I \leq TI_u$, different areas, such as background or | + | Using a thresholding, $TS_l \leq s(p) \leq TS_u \wedge TI_l \leq I \leq TI_u$, different areas, such as background or pathogen, can be selected. |
For the empirical choice of thresholds it can be argued that these are hand tailored to the specific case. | For the empirical choice of thresholds it can be argued that these are hand tailored to the specific case. | ||
| Line 231: | Line 229: | ||
A proper theoretical evaluation is emphasized, however probably is not the aim of the iGEM competition. | A proper theoretical evaluation is emphasized, however probably is not the aim of the iGEM competition. | ||
| - | Finally selecting for the red region, this delivers the location of possible | + | Finally selecting for the red region, this delivers the location of possible pathogens. |
Since the size of the agar chips is variable but fixed, a quantitative analysis can be performed by counting pixels for instance. | Since the size of the agar chips is variable but fixed, a quantitative analysis can be performed by counting pixels for instance. | ||
Revision as of 09:03, 17 October 2014
|
|
|
|
|
|
 "
"