Team:IvyTech SouthBend IN/progress
From 2014.igem.org
(Difference between revisions)
| Line 102: | Line 102: | ||
<ul> | <ul> | ||
<li>This week our team had to split up. Two of our members-Kevin and Lyuda- went along with our adviser Professor Twaddle to Posters on the Hill for CCURI (Community College Undergraduate Research Initiative) in Washington D.C. While they were away, Karinne worked on subculturing into fresh broth so that we can make sure to have clean cell lines to work with.</li><br /> | <li>This week our team had to split up. Two of our members-Kevin and Lyuda- went along with our adviser Professor Twaddle to Posters on the Hill for CCURI (Community College Undergraduate Research Initiative) in Washington D.C. While they were away, Karinne worked on subculturing into fresh broth so that we can make sure to have clean cell lines to work with.</li><br /> | ||
| - | <li>Also, after an exchange of emails with the Carnegie Melon iGEM team we realized we would be able to help test a beta kit of theirs that make the DNA of wheat germ visible after only a few simple steps. It was easy to use and easy to understand and we thank the team profusely for the fun experience | + | <li>Also, after an exchange of emails with the Carnegie Melon iGEM team we realized we would be able to help test a beta kit of theirs that make the DNA of wheat germ visible after only a few simple steps. It was easy to use and easy to understand and we thank the team profusely for the fun experience of this experiment!</li> |
| - | <li>We also started delving into the nanotech side of our project by running a protocol of various monolayers to test adhesion for the most favorable surface for biofilm adhesion. We are doing this to make a more optimal environment for the | + | <li>We also started delving into the nanotech side of our project by running a protocol of various monolayers to test adhesion for the most favorable surface for biofilm adhesion. We are doing this to make a more optimal environment for the lyophilized cells that will be kept in the chamber of our device until a sample is introduced. 2% solution in acetone was used. After monolayers were added to slides they were incubated while being soaked in broth cultures of Top 10 E. coli cells. (3-Aminopropyl)triethoxysilane (APTES) was found to have the most cells attached when counting results under a microscope. </li> |
</ul> | </ul> | ||
<!-- column 1 end --> | <!-- column 1 end --> | ||
| Line 164: | Line 164: | ||
<div id="memo2g"><a name="9713">Week</a> of 9/7/14 through 9/13/14</div> | <div id="memo2g"><a name="9713">Week</a> of 9/7/14 through 9/13/14</div> | ||
<ul> | <ul> | ||
| - | <li>Continuing in our goal towards collaboration with other iGEM teams, we came into contact with the Cornell iGEM team. They are working on detecting heavy metal pollution in water. To aid them in their research, we sent them two 50cc water samples from local sources (including the Saint Joseph river and | + | <li>Continuing in our goal towards collaboration with other iGEM teams, we came into contact with the Cornell iGEM team. They are working on detecting heavy metal pollution in water. To aid them in their research, we sent them two 50cc water samples from local sources (including the Saint Joseph river and Potato Creek.) Hopefully this helped them out!</li><br /> |
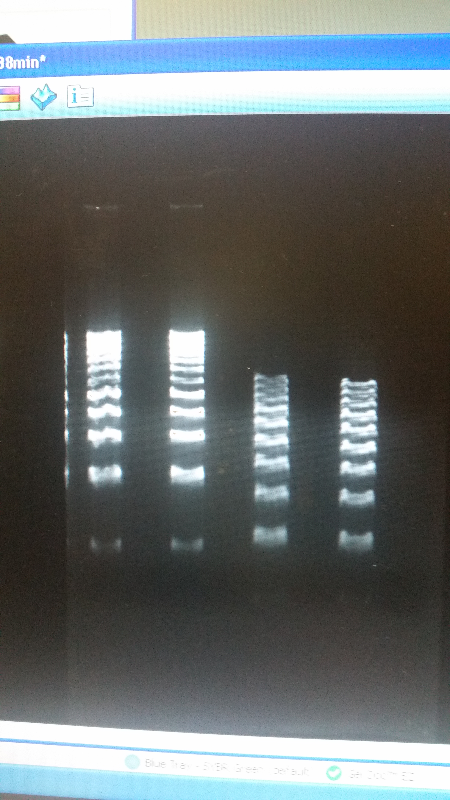
<li>As well as our journey towards working with other teams, this week we reverted back to running an electrophoresis gel for our parts. With the help of a new addition to our advisers-Shaunasee Kocen-we found newer materials, and began using different sized DNA ladders to compare. After a few more tries, we were able to get our DNA bands looking great, and we are ready to send our parts into the iGEM registry.</li><br /> | <li>As well as our journey towards working with other teams, this week we reverted back to running an electrophoresis gel for our parts. With the help of a new addition to our advisers-Shaunasee Kocen-we found newer materials, and began using different sized DNA ladders to compare. After a few more tries, we were able to get our DNA bands looking great, and we are ready to send our parts into the iGEM registry.</li><br /> | ||
<li>Left image is our part, K1477014. Right image is our part, K1477030</li> | <li>Left image is our part, K1477014. Right image is our part, K1477030</li> | ||
| Line 190: | Line 190: | ||
<li>To begin, we exchanged emails with the ETH Zurich team. They asked us to complete surveys as well as send them to friends and family members.</li><br /> | <li>To begin, we exchanged emails with the ETH Zurich team. They asked us to complete surveys as well as send them to friends and family members.</li><br /> | ||
<li>Altogether, we were able to fill out over 30 surveys.</li><br /> | <li>Altogether, we were able to fill out over 30 surveys.</li><br /> | ||
| - | <li>However, we were not able to reach our goal of 50 which was disappointing. Still, the Zurich team | + | <li>However, we were not able to reach our goal of 50 which was disappointing. Still, the Zurich team graciously rewarded our efforts with a team badge for our wiki in appreciation of completing the surveys.</li> |
</ul> | </ul> | ||
<!-- column 1 end --> | <!-- column 1 end --> | ||
| Line 266: | Line 266: | ||
<ul> | <ul> | ||
<li>During this week we restocked our supplies for broth and agar growth media, as we were running very low. </li><br /> | <li>During this week we restocked our supplies for broth and agar growth media, as we were running very low. </li><br /> | ||
| - | <li>One of our advisers suggested we change the buffer we were using for our gel | + | <li>One of our advisers suggested we change the buffer we were using for our gel electrophoresis, and that when preparing the DNA mixture to insert into the gel, we should use buffer instead of DDI water, since this is what is present when the gel is running.</li><br /> |
<li>However, even after changing the buffer, we did not experience any better results. We tried a few times. First, the DNA of our parts did not show up at all. And again, our DNA ladder did not show up correctly.</li> | <li>However, even after changing the buffer, we did not experience any better results. We tried a few times. First, the DNA of our parts did not show up at all. And again, our DNA ladder did not show up correctly.</li> | ||
</ul> | </ul> | ||
Revision as of 17:46, 13 October 2014
Week of 8/3/14 through 8/9/14
- During this week, the team realized that the contamination was coming from our stock of cells that was being kept in the -80C freezer. We have no idea how they became contaminated, seeing as continued to use the line of cells we froze for a week after we froze them, but before any contamination occurred. Nonetheless, one of our advisers has procured a new stock of top 10 E. coli cells for us to use.
- After unsuccessful runs of electrophoresis gels, we have run out of DNA to try to measure. So, we took some time out this week to streak, grow, and purify lines of our K1477014 and K1477030 parts on both pSB1C3 and pSB1AT3 backbones.
Week of 7/13/14 through 7/19/14
- This week we worked towards switching our part's backbone from pSB1AT3 to pSB1C3. We had some difficulty doing this. The first attempt was fruitless, but on our second attempt we had success and we now have a stock of our part on a chlor-resistant backbone.
- Unfortunately, We no longer have any stock of our original part. So, now we are again following the Biolabs Biobrick Assembly Manual to reconstruct our parts K1477014 and K1477030. Our first try yielded nothing.
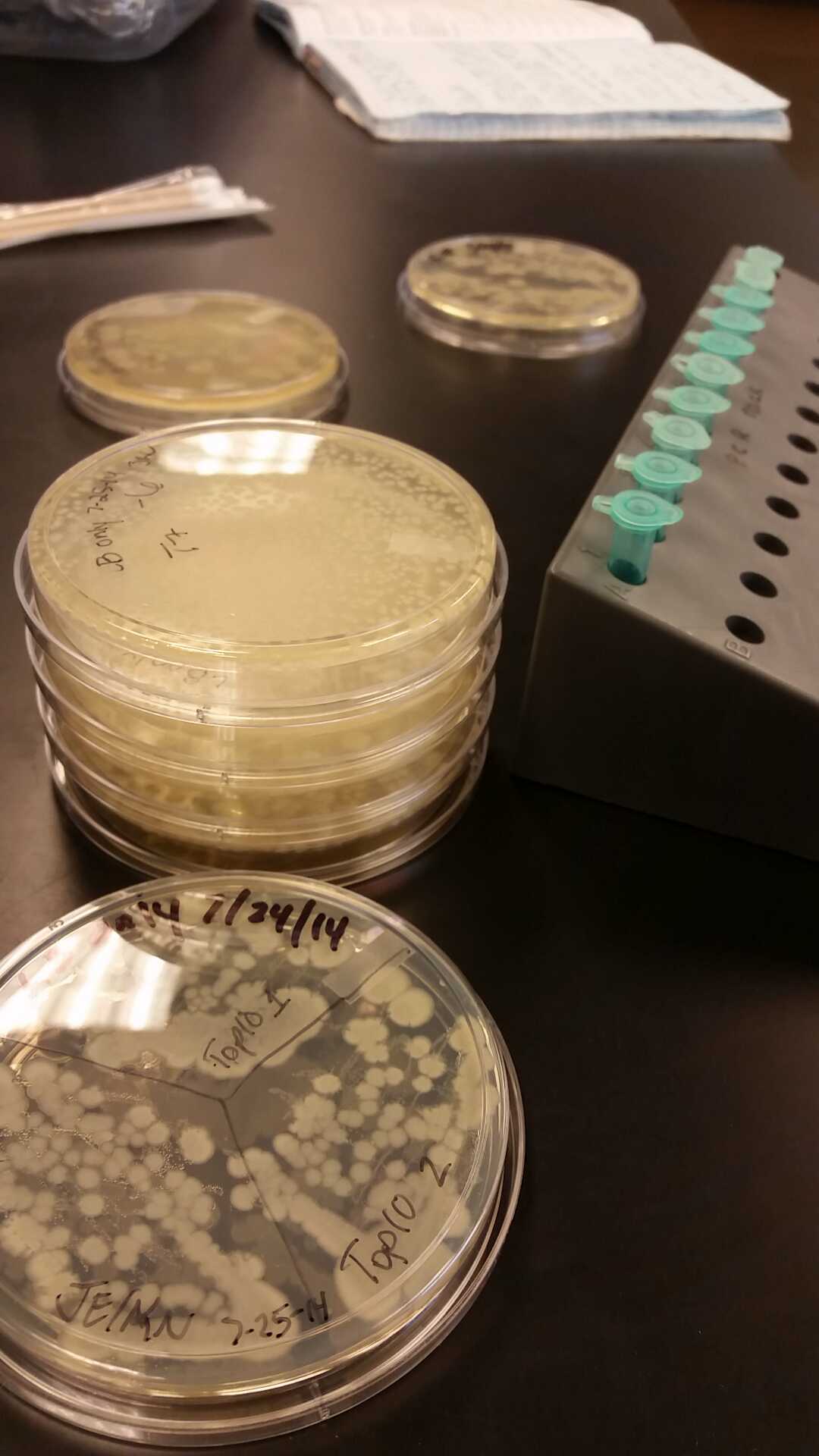
- Our second run through of the protocol resulted in the in the growth of red colonies on chlor plates after electroportion of our part into Top 10 cells. However, we need white colonies to grow because this will point towards the rfp being repressed when the three parts come together. Red colonies are a sign that we did not have a successful assembly of our parts.
Week of 5/25/14 through 5/31/14
- We transformed Top 10 cells through electroporation with registry parts K112806 (Endolysin,) I732020 (LacZoperon Mutant,) J23102 (p-con) and I712074 (T7 promoter.)
- After streaking on plates, and leaving them in the incubator for a day, there was growth on all plates, showing that a successful transformation occured.
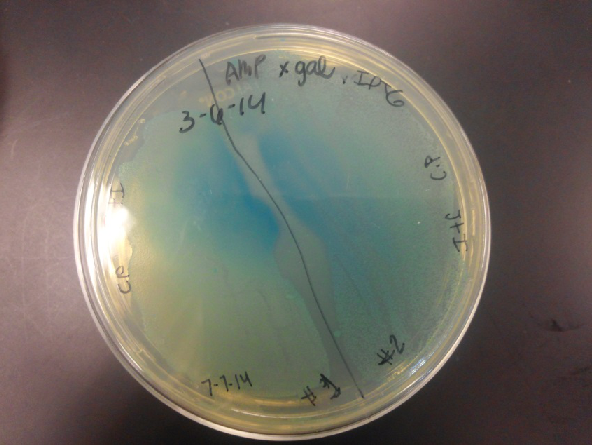
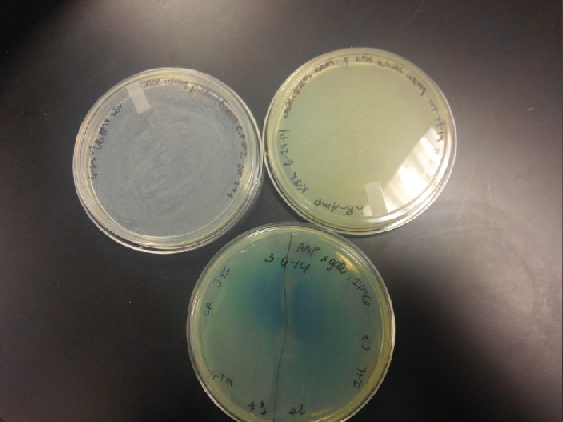
- Plates containing J23102 were red-another sign of success. Unfortunately, it looks like contamination has arisen as well, and is spreading on plates with I712074 and K112806 growth.
- The contamination persisted all week, but we were able to get plates with transformed parts that contained no contamination growth.
- We then ran a DNA purification protocol of J23102 and I732020 to get them ready for a biobrick assembly protocol.
 "
"