Team:Heidelberg
From 2014.igem.org
(Difference between revisions)
| Line 85: | Line 85: | ||
<div class="row"> | <div class="row"> | ||
| - | <div class="jumbotron slide | + | <div class="jumbotron slide epic-background" style="color:white;"> |
| - | + | <div class="col-lg-4 col-md-4 col-xs-12" style="height: 410px; position: relative"> | |
| - | + | <div id="circ-box" class="descr-box"> | |
| - | + | <a href="/Team:Heidelberg/Toolbox/Circularization"> | |
| + | <img src="/wiki/images/5/58/Heidelberg_Toolbox_Circularization.png" id="circ-icon" class="toolbox-icon"> CIRCULARIZATION | ||
| + | </a> | ||
</div> | </div> | ||
| - | <div class=" | + | <div id="oligo-box" class="descr-box"> |
| - | < | + | <a href="/Team:Heidelberg/Toolbox/Oligomerization"> |
| - | + | <img src="/wiki/images/4/40/Oligomerization.png" id="oligo-icon" class="toolbox-icon"> OLIGOMERIZATION | |
| - | + | </a> | |
</div> | </div> | ||
| - | <div class=" | + | <div id="fusion-box" class="descr-box"> |
| - | < | + | <a href="/Team:Heidelberg/Toolbox/Fusion"> |
| + | <img src="/wiki/images/8/87/Heidelberg_Toolbox_Fusion.png" id="fusion-icon" class="toolbox-icon"> FUSION | ||
| + | </a> | ||
</div> | </div> | ||
| - | <div | + | <div id="onoff-box" class="descr-box"> |
| - | + | <a href="/Team:Heidelberg/Toolbox/OnOff"> | |
| - | + | <img src="/wiki/images/c/c2/Heidelberg_Toolbox_On-Off.png" id="onoff-icon" class="toolbox-icon"> ON/OFF | |
| - | + | </a> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | <img | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | </ | + | |
| - | + | ||
</div> | </div> | ||
| - | <div class=" | + | <div id="purification-box" class="descr-box"> |
| - | + | <a href="/Team:Heidelberg/Toolbox/Purification"> | |
| - | + | <img src="/wiki/images/0/04/Heidelberg_Toolbox_Purification.png" id="purification-icon" class="toolbox-icon"> PURIFICATION | |
| - | + | </a> | |
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
| + | <div class="col-lg-8 col-md-8 col-xs-12"> | ||
| + | <h1 style="text-align: right; font-size: 3em;" >... and show you the WORLD of <br /> | ||
| + | <span class="red-text">post-translational MODIFCATION</span> | ||
| + | </h1> | ||
| + | <p>The iGEM Team Heidelberg has developed an intein toolbox for the iGEM community to easily modify your protein in a standarized method.</p> | ||
| + | <p>Our toolbox contains several tools which are supergeil. Here you can find out more about our Toolbox.</p> | ||
| + | <p>In addition to that all tools are inducible by light. Using the LOV system we built a solid method for regulation of the intein trans-splicing reaction our toolbox consiting on. Click here to get more informations about Induction.</p> | ||
| + | </div> | ||
| + | <div class="clearfix"></div> | ||
<div class="arrow arrow-left"></div> | <div class="arrow arrow-left"></div> | ||
</div> | </div> | ||
</div> | </div> | ||
| - | |||
<div style="position:relative;" class="row"> | <div style="position:relative;" class="row"> | ||
<div class="jumbotron slide epic-background" style="color:white;"> | <div class="jumbotron slide epic-background" style="color:white;"> | ||
| Line 174: | Line 170: | ||
</div> | </div> | ||
</div> | </div> | ||
| - | |||
| - | |||
<div class="row"> | <div class="row"> | ||
| - | <div class="jumbotron slide | + | <div class="jumbotron slide dark-grey" style="color:white;"> |
| - | <div class="col- | + | <div> |
| - | + | <div class="col-md-4 col-md-push-8 col-md-offset-0 col-xs-offset-3 col-xs-6" style="z-index:5;"> | |
| - | + | <img class="img-responsive" src="/wiki/images/d/df/Heidelberg_Lysozyme.png" /> | |
| - | + | ||
| - | + | ||
</div> | </div> | ||
| - | <div | + | <div class="col-md-8 col-md-pull-4 col-xs-12"> |
| - | < | + | <h1 style="text-align: left;" ><span style="font-size:1.3em;">LINK it!</span></h1> |
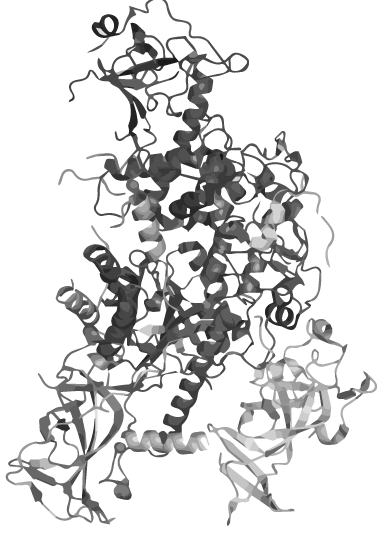
| - | + | <p>Could every protein becomes heat stable by circularization, even if it´s the most complex of all?</p> | |
| - | </a> | + | <p>Circularization is a narrow path between gaining heat-stability and loosing function due to deformation. We developed a linker software, which predict the perfect linker depending on the folding structure of every protein.</p> |
</div> | </div> | ||
| - | <div | + | <div class="col-lg-12"> |
| - | < | + | <p>In an extensive linker screening our software was improved and calibrated using the lambda phage lysozyme.</p> |
| - | + | ||
| - | + | ||
</div> | </div> | ||
| - | <div | + | <div class="clearfix"></div> |
| - | < | + | </div> |
| - | <img src="/wiki/images/ | + | <div class="arrow arrow-left"></div> |
| - | </ | + | </div> |
| + | </div> | ||
| + | <div class="row"> | ||
| + | <div class="jumbotron slide grey"> | ||
| + | <div> | ||
| + | <div> | ||
| + | <div class="col-md-3 col-md-offset-0 col-xs-6 col-xs-offset-3" style="z-index:5;"> | ||
| + | <img class="img-responsive" src="/wiki/images/e/e5/Heidelberg_Frontpage_igemathome.png" /> | ||
| + | </div> | ||
| + | <div class="col-md-9 col-xs-12"> | ||
| + | <h1 style="text-align: left; color:#DE4230;" ><span style="font-size:1.5em; font-weights:bold;">CALCULATE it!</span></h1> | ||
| + | </div> | ||
| + | <div class="clearfix"></div> | ||
</div> | </div> | ||
| - | <div | + | <div class="col-md-8 col-xs-12"> |
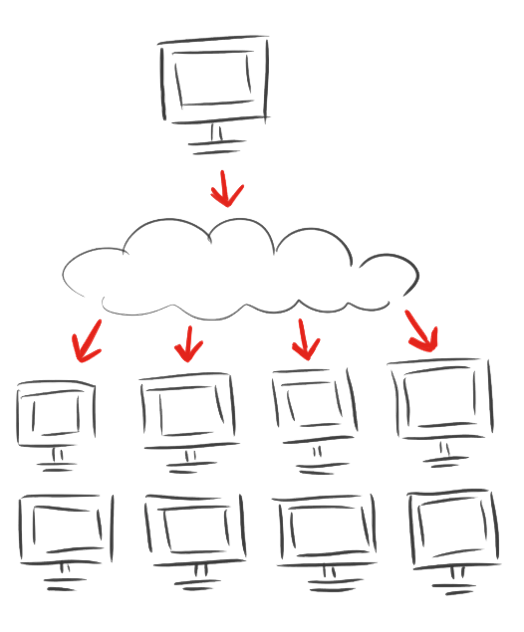
| - | + | <p><br/>After calculating for eleven days and the breakdown of both computational and mental power we decided to spread the modeling of the linkers.</p> | |
| - | + | <p>The iGEM Team Heidelberg developed <a href="/Team:Heidelberg/Software/igemathome">iGEM@home</a>, a software to divide extensive computing task into many packages and to distribute them to many computers. Now over 1,000 volunteers are calculating for us when their computers are idle.</p> | |
| - | + | <p>As a new tool for the iGEM community this system enables every student team to archieve their modeling without access to big server farms.</p> | |
</div> | </div> | ||
| + | <div class="col-md-4 col-md-offset-0 col-sm-offset-3 col-sm-6 col-xs-offset-2 col-xs-8"> | ||
| + | <img style="position:relative; top: -35px;" class="img-responsive" src="/wiki/images/5/52/Heidelberg_Frontpage_igemathome_cloud.png" /> | ||
| + | </div> | ||
| + | <div class="clearfix"></div> | ||
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<div class="arrow arrow-left"></div> | <div class="arrow arrow-left"></div> | ||
</div> | </div> | ||
Revision as of 15:07, 13 October 2014
 "
"


 CIRCULARIZATION
CIRCULARIZATION
 OLIGOMERIZATION
OLIGOMERIZATION
 FUSION
FUSION
 ON/OFF
ON/OFF
 PURIFICATION
PURIFICATION