Team:Aachen/Project/2D Biosensor
From 2014.igem.org
(→Principle of Operation) |
|||
| Line 148: | Line 148: | ||
=== Induction of the Sensor Chips === | === Induction of the Sensor Chips === | ||
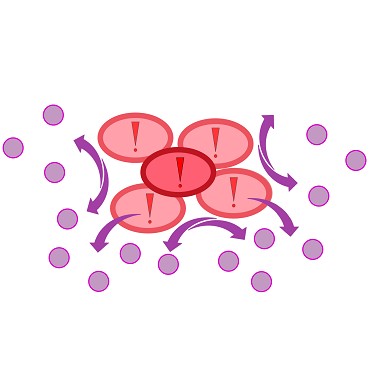
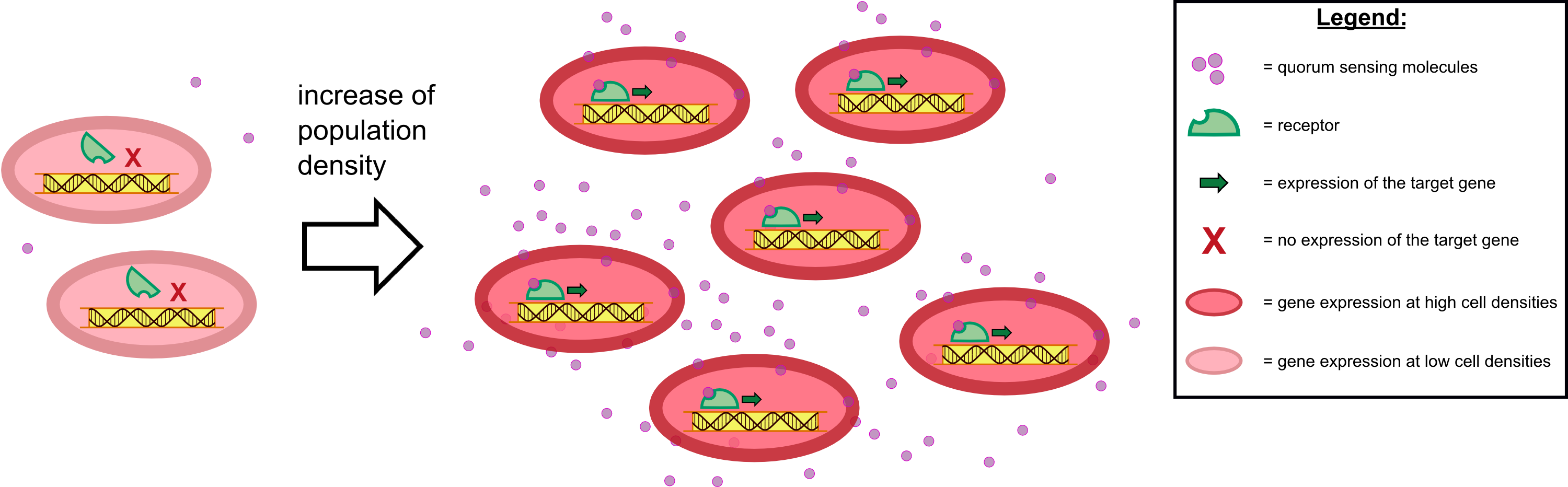
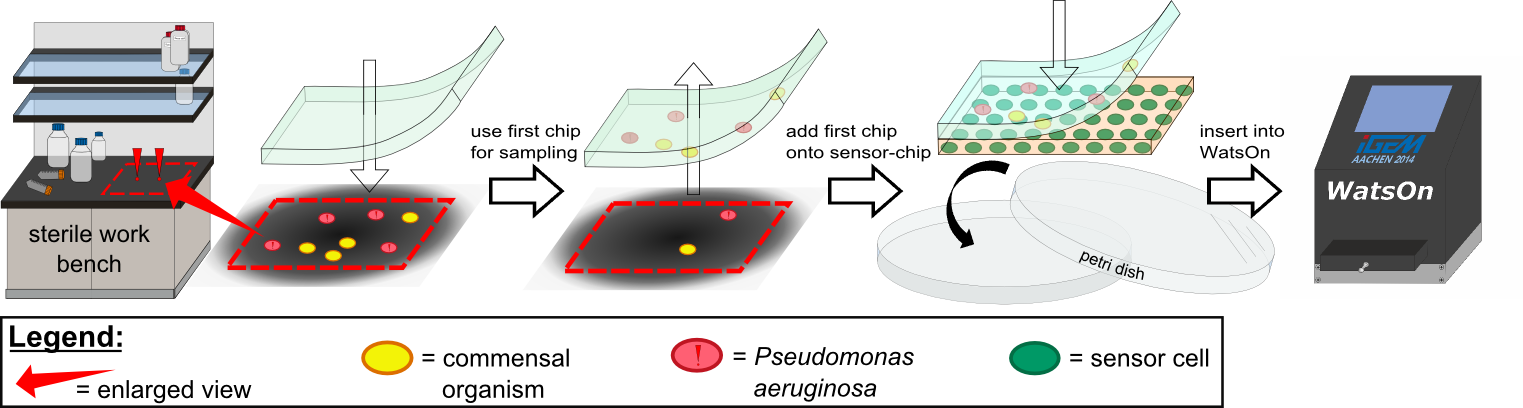
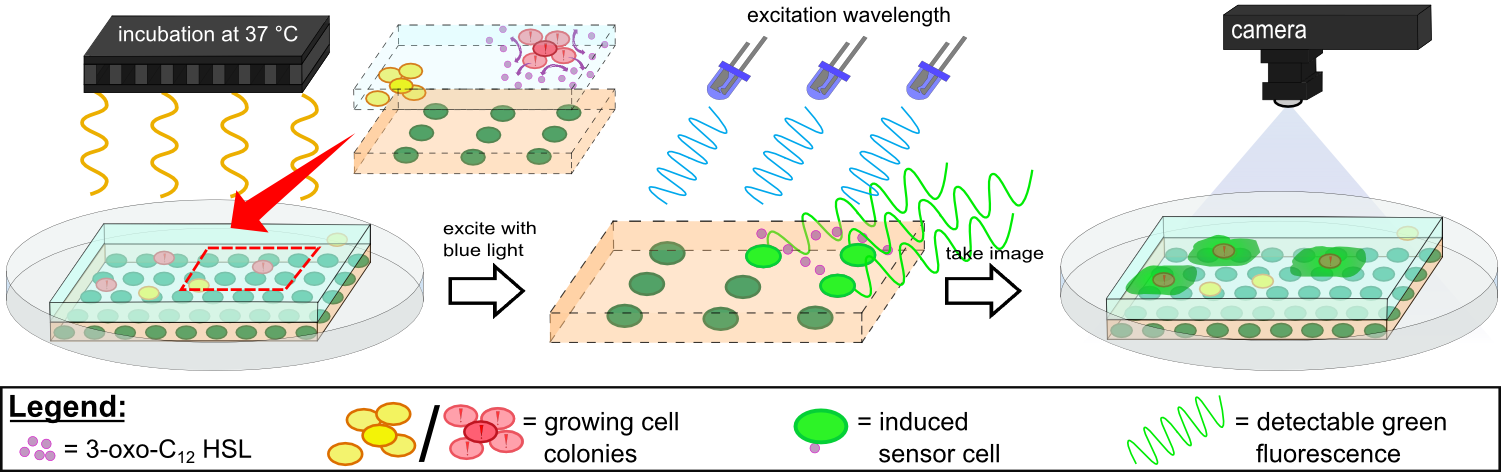
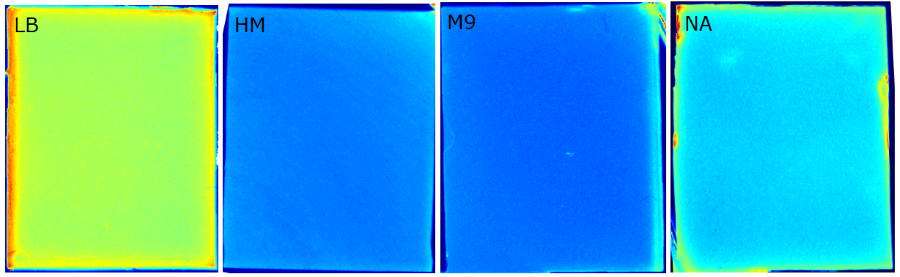
To test our molecular constructs, we simulated the presence of ''P. aeruginosa'' by using IPTG or 3-oxo-C<sub>12</sub>-HSL. Initial experiments showed that diffusion of the inducers hinder the formation of distinct fluorescent spots. Through this set of experiments we determined that the best compromise between diffusion and spot intensity is an induction volume of 2.0 µL for IPTG and 0.2 µL for HSL. Furthermore, detection of growing ''P. aeruginosa'' based on secreted HSLs was possible using the [http://parts.igem.org/Part:BBa_K131026 K131026] construct. The experiments for optimizing the induction of our sensor chips are described in more detail in the [https://2014.igem.org/Team:Aachen/Project/2D_Biosensor#biosensorachievements Achievements] section. | To test our molecular constructs, we simulated the presence of ''P. aeruginosa'' by using IPTG or 3-oxo-C<sub>12</sub>-HSL. Initial experiments showed that diffusion of the inducers hinder the formation of distinct fluorescent spots. Through this set of experiments we determined that the best compromise between diffusion and spot intensity is an induction volume of 2.0 µL for IPTG and 0.2 µL for HSL. Furthermore, detection of growing ''P. aeruginosa'' based on secreted HSLs was possible using the [http://parts.igem.org/Part:BBa_K131026 K131026] construct. The experiments for optimizing the induction of our sensor chips are described in more detail in the [https://2014.igem.org/Team:Aachen/Project/2D_Biosensor#biosensorachievements Achievements] section. | ||
| + | === Negative Control === | ||
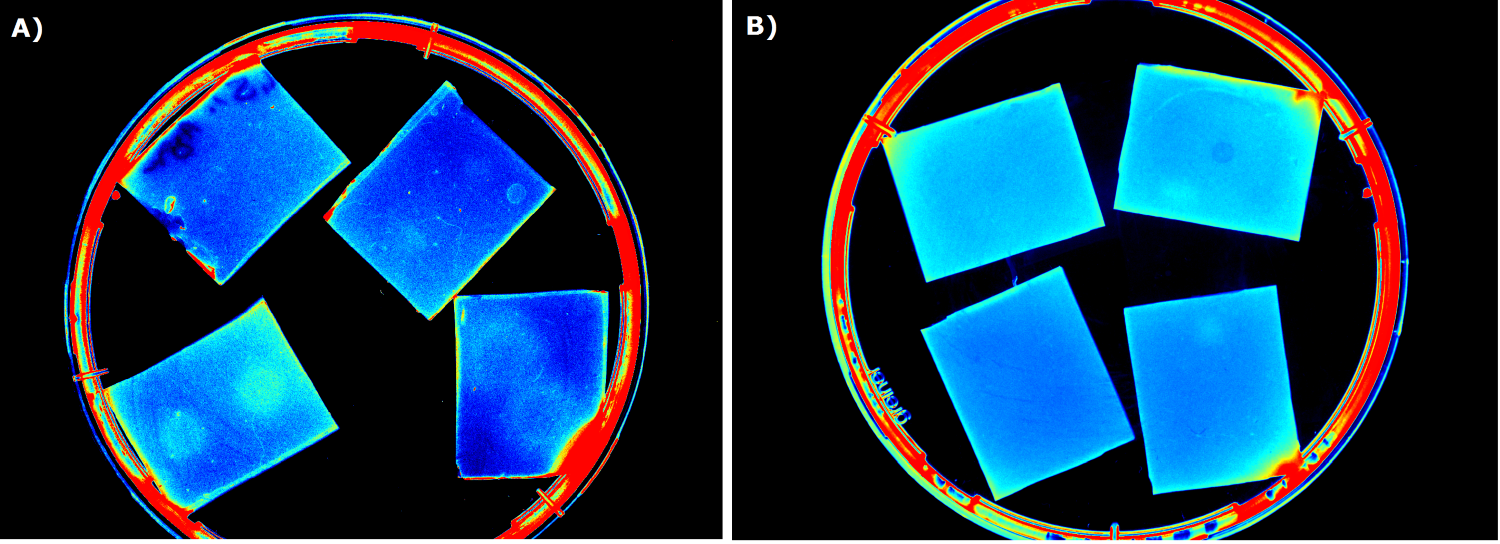
| + | To ensure that the fluorescence signal resulted from the sensor construct and not from the medium or ''E. coli'' cells themselves, [http://parts.igem.org/Part:BBa_B0015 B0015] in NEB was used as negative control during sensor chip induction with IPTG, HSL and ''P. aeruginosa'' (Negativ control, displayed below). | ||
| + | {{Team:Aachen/Figure|Aachen_B0015_IPTG_HSL_Pseudomonas.png|title=Negativ control |subtitle=B0015 in NEB as negativ control induced with A) 0.2 µl of 100 mM IPTG, image taken after 2.5 h; B) 0.2 µl of 500 µg/ml HSL (3-oxo-C12), image after 2.5 h; C) with 5 spots of ''Pseudomonas aeruginosa'' on the left and one big spot on the right, image taken after 2 h|width=900px}} | ||
{{Team:Aachen/BlockSeparator}} | {{Team:Aachen/BlockSeparator}} | ||
| Line 157: | Line 160: | ||
<span class="anchor" id="biosensorachievements"></span> | <span class="anchor" id="biosensorachievements"></span> | ||
| - | We developed and optimized a 2D biosensor, which is able to detect IPTG, 3-oxo-C{{sub|12}}-HSL and living cells of the pathogen ''Pseudomonas aeruginosa''. | + | We developed and optimized a 2D biosensor, which is able to detect IPTG, 3-oxo-C{{sub|12}}-HSL and living cells of the pathogen ''Pseudomonas aeruginosa''. |
| - | + | ||
| - | + | ||
=== Testing our Sensor Chips in a Plate Reader === | === Testing our Sensor Chips in a Plate Reader === | ||
Revision as of 01:35, 18 October 2014
|
|
|
|
|
|
 "
"