Introduction
In the native DCM-degrading bacterium the following genetic construct exists:
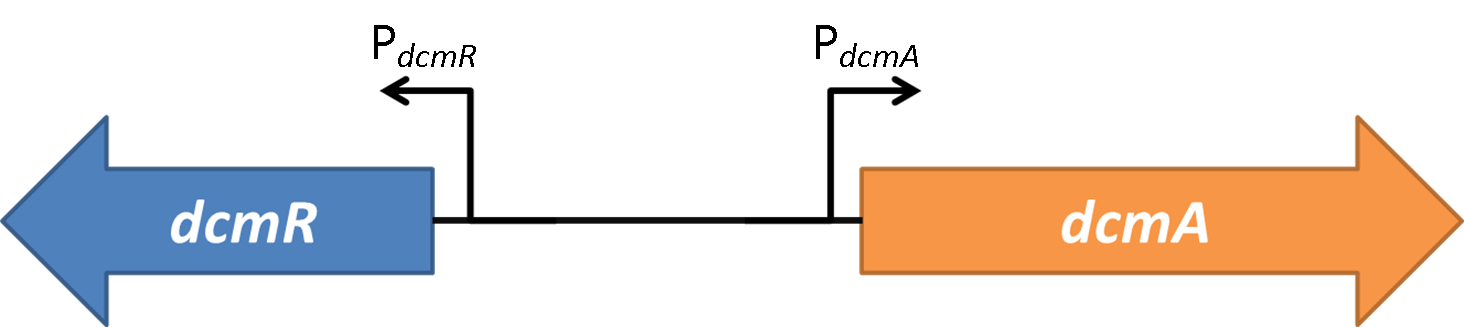
Oxford iGEM has isolated this intergenic region that contains a bidirectional promoter. We have termed these "PdcmA" acting towards the right in the direction of the gene encoding DcmA, and "PdcmR" acting towards the left in the direction of the gene encoding DcmR.
The literature claims DcmR to be a regulatory protein acting to control the expression genes encoding DcmA and DcmR. Little is known about the different parameters in each direction.
We have observed and characterised the effect of DcmR regulating both promoters; in both directions under the influence of DcmR.
This is a new tool for molecular biologists wishing to manipulate stoichiometries of genetic products in the same construct.
We have also characterised PdcmA repression as a double repression system with DCM modulating DcmR activity resulting in derepression of genes downstream of PdcmA. This DCM sensitivity is not observed in the PdcmR direction.
Wetlab Results:
Is DcmR a repressor or an activator at PdcmA/PdcmR promoters?

Conclusion: DcmR is a repressor of both the PdcmA and PdcmR promoter.
DcmR at the bidirectional promoter - A new tool for molecular biologists

We can see from these results that at both growth stages the relative expression is weighted towards PdcmR. An interesting result is that in the presence of DcmR this ratio is reduced such that expression through PdcmR is relatively increased while expression through PdcmA is relatively decreased. This result is observed as a decrease in the PdcmA/PdcmR expression ratio. From our results this appears more statistically relevant in stationary phase. The cut off for exponential/stationary phase was set at 600 minutes as seen in the above graph.
Through analysing this new bidirectional promoter we have discovered a new tool for molecular biologists. Our new intergenic region is able to modulate different stoichiometries between genes inserted either side of it in response to the presence or absence of DcmR (available in our BioBrick BBa_K1446003). The relative stoichiometry (PdcmA/PdcmR activation) of ~0.9 being reduced to ~0.3 upon DcmR addition.
DCM interacting with DcmR - the basis for our biosensor
For the PdcmA promoter both with and without DcmR:

For the PdcmR promoter both with and without DcmR:

From these results we can see that in the direction of PdcmA DCM has a modulating effect of relieving DcmR repression thus DCM acts through depression. Interestingly, DCM appears to decrease sfGFP fluorescence in the absence of DcmR.
In the direction of PdcmR DCM appears to have no significant effect on the expression of sfGFP. Therefore in the native system we suggest that DCM has a modulating effect through depression of dcmA expression. Additionally DCM shows no effect on the auto regulation of dcmR expression.
In the context of our biosensor this means that DCM is capable of modulating expression of sfGFP through interaction through the PdcmA repressor DcmR.
Ultimately we can conclude that DCM acts to encourage expression of sfGFP through derepression: consistent with our double repression model.
Discussion:
The results displayed here demonstrate the repression nature of DcmR on PdcmA and PdcmR bidirectional promoter. The modulation by DCM on the PdcmA promoter we have observed offers a constructive route towards a Dichloromethane biosensor. In this way we are able to use a reporter gene downstream of PdcmA to respond to the concentration of DCM.
Additionally the nature of the bidirectional promoter offers a uniquely useful tool to molecular biologists. The relative stoichiometry of two genes inserted either side of the bidirectional promoter can be can be modulated through the addition of DcmR (BBa_K1446003 - Oxford iGEM). This can be utilised easily for investigations into the effect of protein stoichiometries between two interacting proteins.
 "
"


 Oxford iGEM has isolated this intergenic region that contains a bidirectional promoter. We have termed these "PdcmA" acting towards the right in the direction of the gene encoding DcmA, and "PdcmR" acting towards the left in the direction of the gene encoding DcmR.
The literature claims DcmR to be a regulatory protein acting to control the expression genes encoding DcmA and DcmR. Little is known about the different parameters in each direction.
Oxford iGEM has isolated this intergenic region that contains a bidirectional promoter. We have termed these "PdcmA" acting towards the right in the direction of the gene encoding DcmA, and "PdcmR" acting towards the left in the direction of the gene encoding DcmR.
The literature claims DcmR to be a regulatory protein acting to control the expression genes encoding DcmA and DcmR. Little is known about the different parameters in each direction.
 Conclusion: DcmR is a repressor of both the PdcmA and PdcmR promoter.
Conclusion: DcmR is a repressor of both the PdcmA and PdcmR promoter.
 We can see from these results that at both growth stages the relative expression is weighted towards PdcmR. An interesting result is that in the presence of DcmR this ratio is reduced such that expression through PdcmR is relatively increased while expression through PdcmA is relatively decreased. This result is observed as a decrease in the PdcmA/PdcmR expression ratio. From our results this appears more statistically relevant in stationary phase. The cut off for exponential/stationary phase was set at 600 minutes as seen in the above graph.
We can see from these results that at both growth stages the relative expression is weighted towards PdcmR. An interesting result is that in the presence of DcmR this ratio is reduced such that expression through PdcmR is relatively increased while expression through PdcmA is relatively decreased. This result is observed as a decrease in the PdcmA/PdcmR expression ratio. From our results this appears more statistically relevant in stationary phase. The cut off for exponential/stationary phase was set at 600 minutes as seen in the above graph.

 From these results we can see that in the direction of PdcmA DCM has a modulating effect of relieving DcmR repression thus DCM acts through depression. Interestingly, DCM appears to decrease sfGFP fluorescence in the absence of DcmR.
In the direction of PdcmR DCM appears to have no significant effect on the expression of sfGFP. Therefore in the native system we suggest that DCM has a modulating effect through depression of dcmA expression. Additionally DCM shows no effect on the auto regulation of dcmR expression.
From these results we can see that in the direction of PdcmA DCM has a modulating effect of relieving DcmR repression thus DCM acts through depression. Interestingly, DCM appears to decrease sfGFP fluorescence in the absence of DcmR.
In the direction of PdcmR DCM appears to have no significant effect on the expression of sfGFP. Therefore in the native system we suggest that DCM has a modulating effect through depression of dcmA expression. Additionally DCM shows no effect on the auto regulation of dcmR expression.
 (A)A western blot showing the expression of a protein at 62 kDa (the calculated molecular weight of DCMA-sfGFP). The existence of some protein product at 0 mM IPTG suggests that the promoter is slightly ‘leaky’ and allows a basal expression level when no inducer is present.
(A)A western blot showing the expression of a protein at 62 kDa (the calculated molecular weight of DCMA-sfGFP). The existence of some protein product at 0 mM IPTG suggests that the promoter is slightly ‘leaky’ and allows a basal expression level when no inducer is present. 






















































