Team:Aachen/Project/Model
From 2014.igem.org
(→Modeling) |
(→Modeling) |
||
| Line 22: | Line 22: | ||
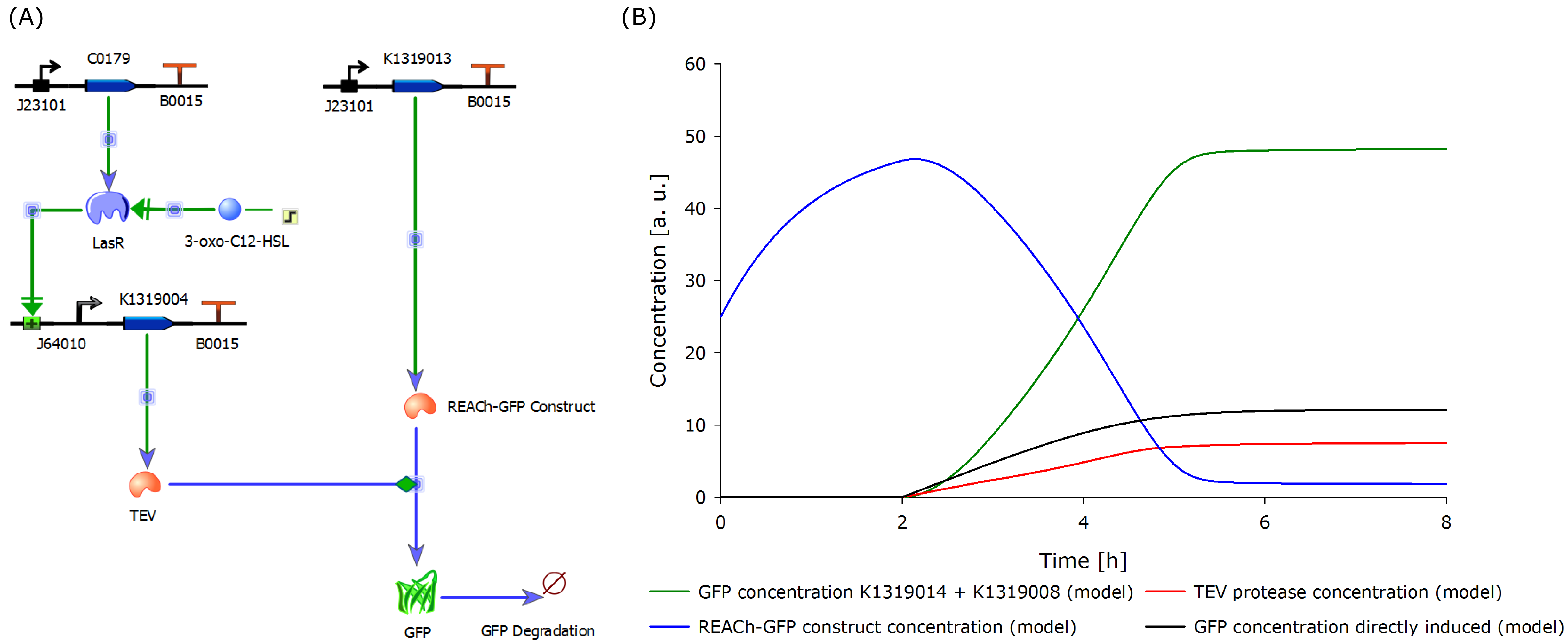
| - | + | In the above figure, we compare the response time of the fluorescence signal between our theoretical system and a traditional biosensor, we included a direct expression of GFP in the same plot. In the results shown above, the strength of the promoter used for the direct GFP expression (traditional approach) is twice as high as the strength of the promoter upstream of the TEV coding sequence in our new approach. Despite a weaker promoter, a '''higher GFP concentration is generated in the model of the novel biosensor''', predicting a quicker response time of our system. | |
The model predicted that our approach should be an improvement over the commonly used direct expression, so we proceeded with the clonings and assembled plasmids to test the system. | The model predicted that our approach should be an improvement over the commonly used direct expression, so we proceeded with the clonings and assembled plasmids to test the system. | ||
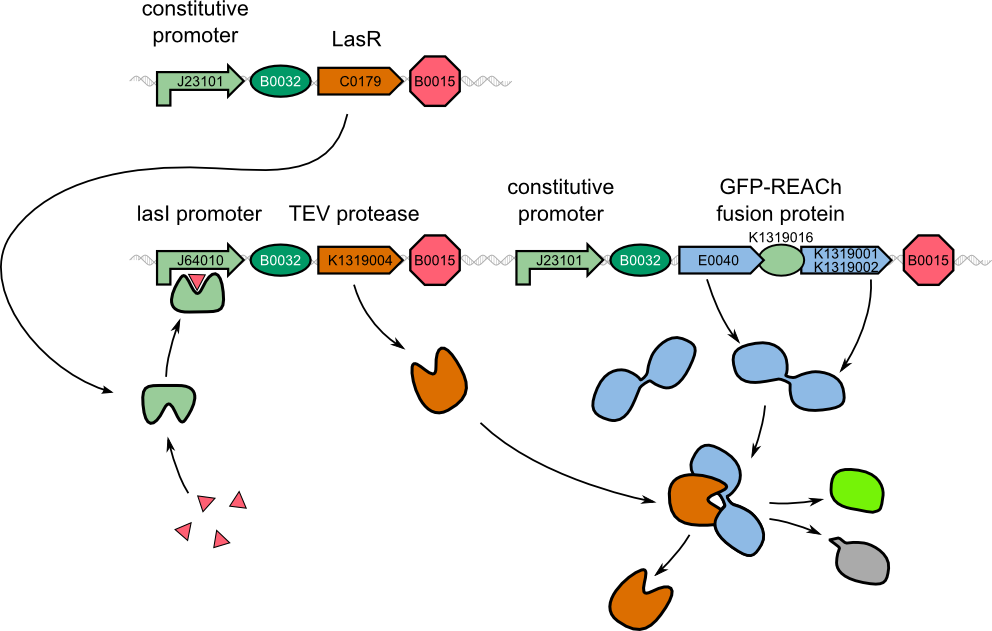
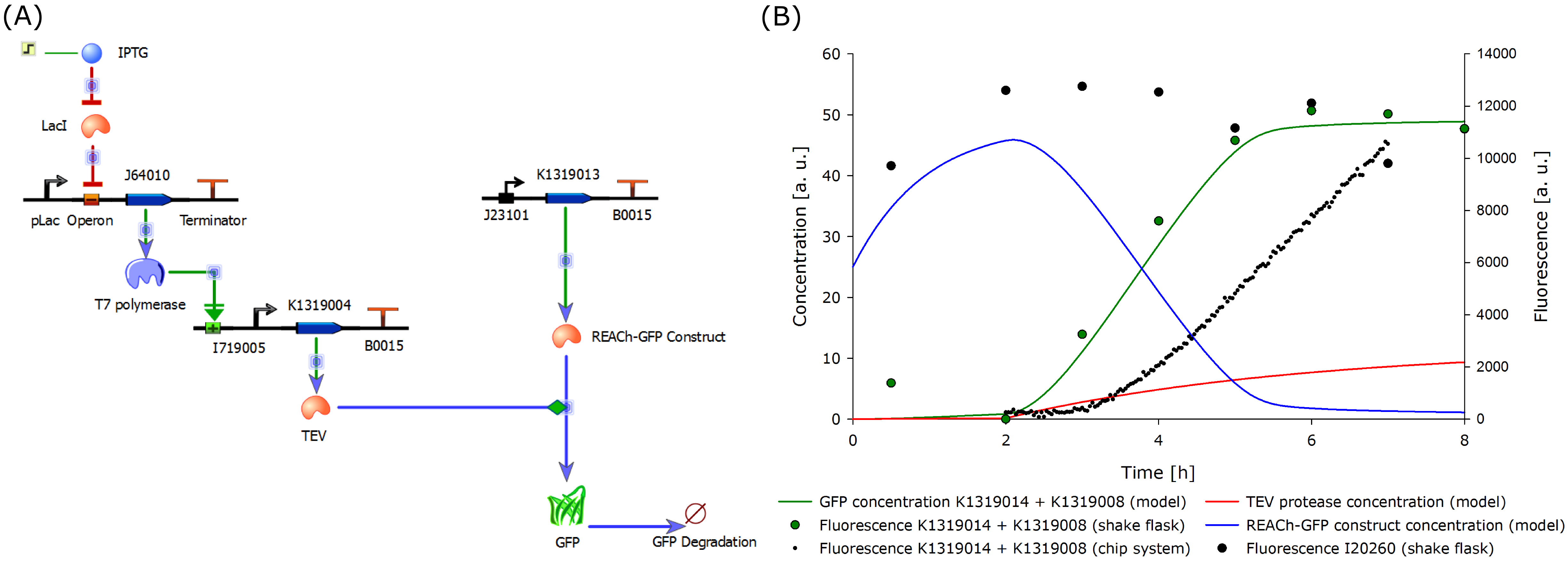
| - | + | Due to the complexity of the quorum sensing circuit, we assembled an IPTG-inducible TEV protease instead of the 3-oxo-C<sub>12</sub>-HSL-inducible version. | |
| - | Although the modeled system and the existing system differ regarding the induction mechanism, the correlation was high. However, since the final 3-oxo-C<sub>12</sub>-HSL inducible construct could not be built in time, a new model was designed according to the existing and functional | + | Although the modeled system and the existing double plasmid system differ regarding the induction mechanism, the correlation was high. However, since the final 3-oxo-C<sub>12</sub>-HSL inducible construct could not be built in time, a new model was designed according to the existing and functional IPTG-inducible system. |
| - | + | ||
<center> | <center> | ||
Revision as of 00:04, 18 October 2014
|
|
 "
"