Team:Aachen/Project/2D Biosensor
From 2014.igem.org
(→Development & Optimization) |
(→Development & Optimization) |
||
| Line 124: | Line 124: | ||
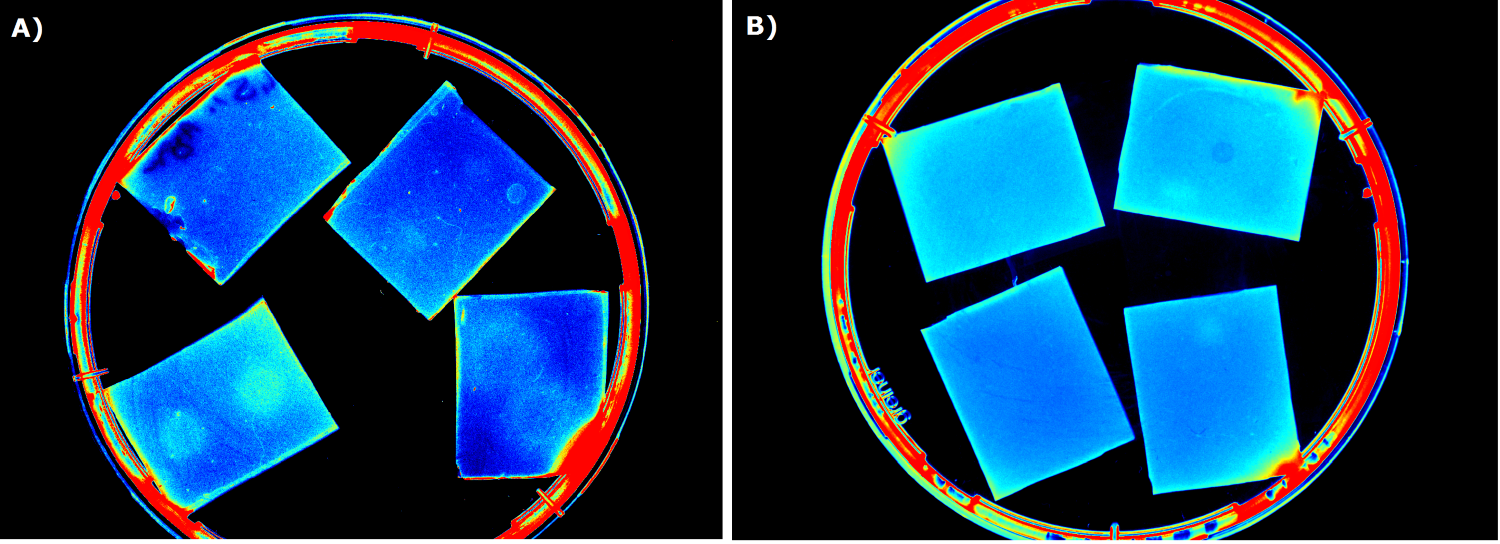
{{Team:Aachen/FigureFloat|Aachen_5days_K131026_neb_tb_1,5h.jpg |title=Testing our chips' shelf-life|subtitle= Chips of [http://parts.igem.org/Part:BBa_K131026 K131026] in NEB were stored 5 days at 4°C. The right chip was induced with 0.2 µL of 500 µg/mL HSL and an image was taken after 1.5 h.|left|width=500px}} | {{Team:Aachen/FigureFloat|Aachen_5days_K131026_neb_tb_1,5h.jpg |title=Testing our chips' shelf-life|subtitle= Chips of [http://parts.igem.org/Part:BBa_K131026 K131026] in NEB were stored 5 days at 4°C. The right chip was induced with 0.2 µL of 500 µg/mL HSL and an image was taken after 1.5 h.|left|width=500px}} | ||
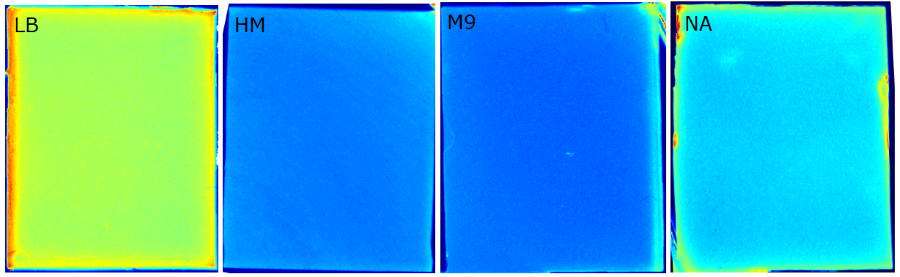
<br> We tested different media (i.e. LB, M9, NA, HM and TB) for the preparation of our sensor chips. The details of media composition can be found in the [https://2014.igem.org/Team:Aachen/Notebook/Protocols Protocols] section. We screened for an optimized media composition that results in minimal background fluorescence and supports cell growth. The resuts of the analysis are presented in the table below. Because of the reduced fluorescence compared to TB medium when using ''WatsOn'' for sensor chip evaluation and because of sufficient cultivation conditions for our ''Cellocks'' '''LB medium was chosen over TB medium for sensor hip manufacturing'''. | <br> We tested different media (i.e. LB, M9, NA, HM and TB) for the preparation of our sensor chips. The details of media composition can be found in the [https://2014.igem.org/Team:Aachen/Notebook/Protocols Protocols] section. We screened for an optimized media composition that results in minimal background fluorescence and supports cell growth. The resuts of the analysis are presented in the table below. Because of the reduced fluorescence compared to TB medium when using ''WatsOn'' for sensor chip evaluation and because of sufficient cultivation conditions for our ''Cellocks'' '''LB medium was chosen over TB medium for sensor hip manufacturing'''. | ||
| - | |||
<center> | <center> | ||
| Line 137: | Line 136: | ||
|} | |} | ||
</center> | </center> | ||
| - | |||
Experiments were conducted to test '''long-time storage''' of the sensor chips at low temperature and by the addition of glycerol. Storage at -20°C resulted in the loss of our sensor cells. Adding 5-10% (v/v) glycerol ensured survival of the sensor cells, but resulted in an expression stop of fluorescence proteins. Hence, we concluded that long time storage of the sensor chips is not possible under the tested conditions. However, it is possible to store 'ready-to-use' sensor chips for 2 days at 4°C when using LB medium and storage for 5 days was possible with chips made from TB medium. | Experiments were conducted to test '''long-time storage''' of the sensor chips at low temperature and by the addition of glycerol. Storage at -20°C resulted in the loss of our sensor cells. Adding 5-10% (v/v) glycerol ensured survival of the sensor cells, but resulted in an expression stop of fluorescence proteins. Hence, we concluded that long time storage of the sensor chips is not possible under the tested conditions. However, it is possible to store 'ready-to-use' sensor chips for 2 days at 4°C when using LB medium and storage for 5 days was possible with chips made from TB medium. | ||
| Line 144: | Line 142: | ||
=== Optimal agarose concentration for sensor chip manufacturing === | === Optimal agarose concentration for sensor chip manufacturing === | ||
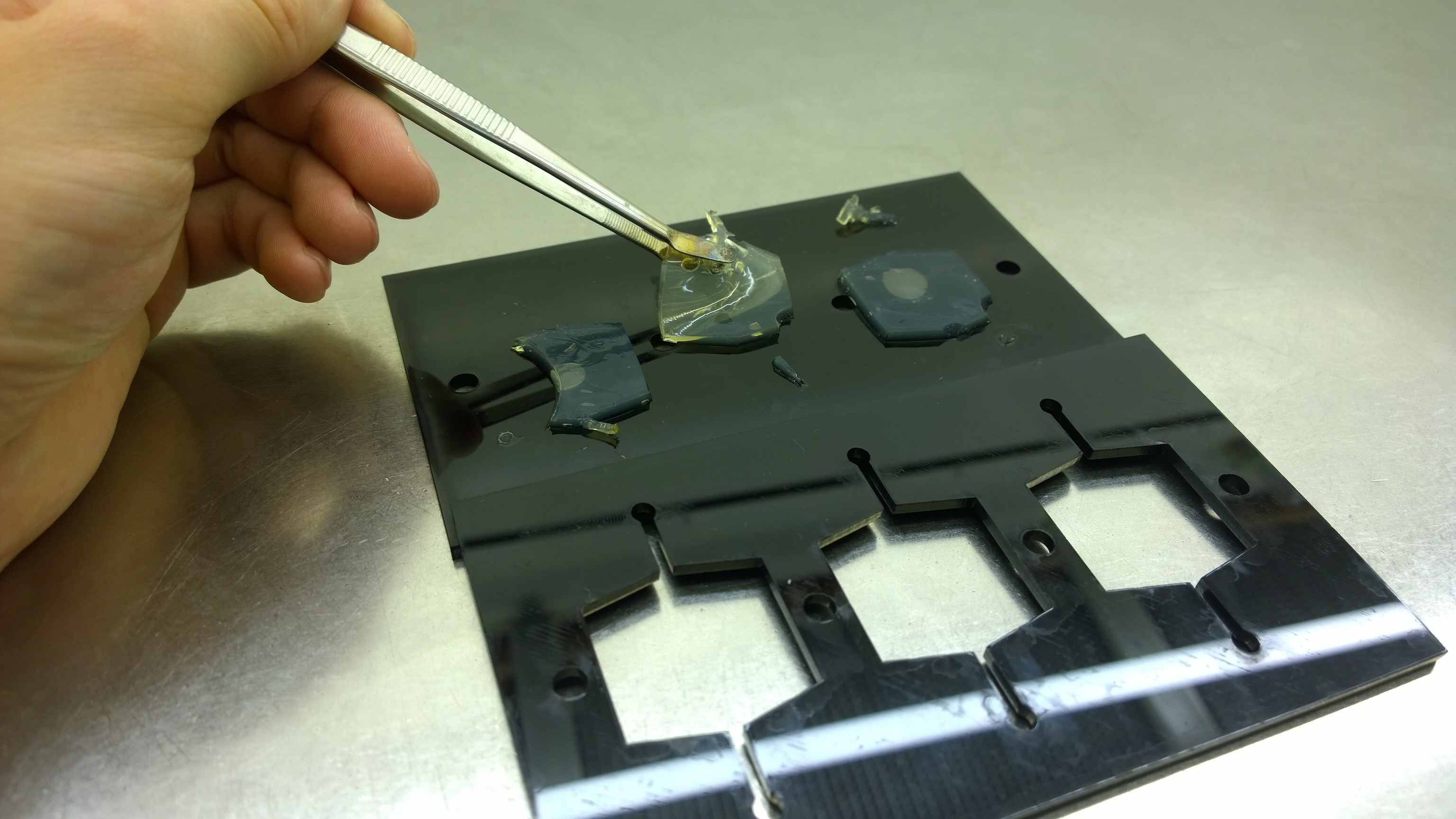
For the sensor chip manufacturing, agarose was preferred over agar, because of a uniform linkage that resulted in a better chip homogeneity. In addition, agarose reduced diffusion of the inducer molecules through the chip. A reduced diffusion is vital to observe distinct fluorescent spots on the sensor chips and thus further optimzation of our 2D biosensor was done using agarose-based chips. | For the sensor chip manufacturing, agarose was preferred over agar, because of a uniform linkage that resulted in a better chip homogeneity. In addition, agarose reduced diffusion of the inducer molecules through the chip. A reduced diffusion is vital to observe distinct fluorescent spots on the sensor chips and thus further optimzation of our 2D biosensor was done using agarose-based chips. | ||
| - | |||
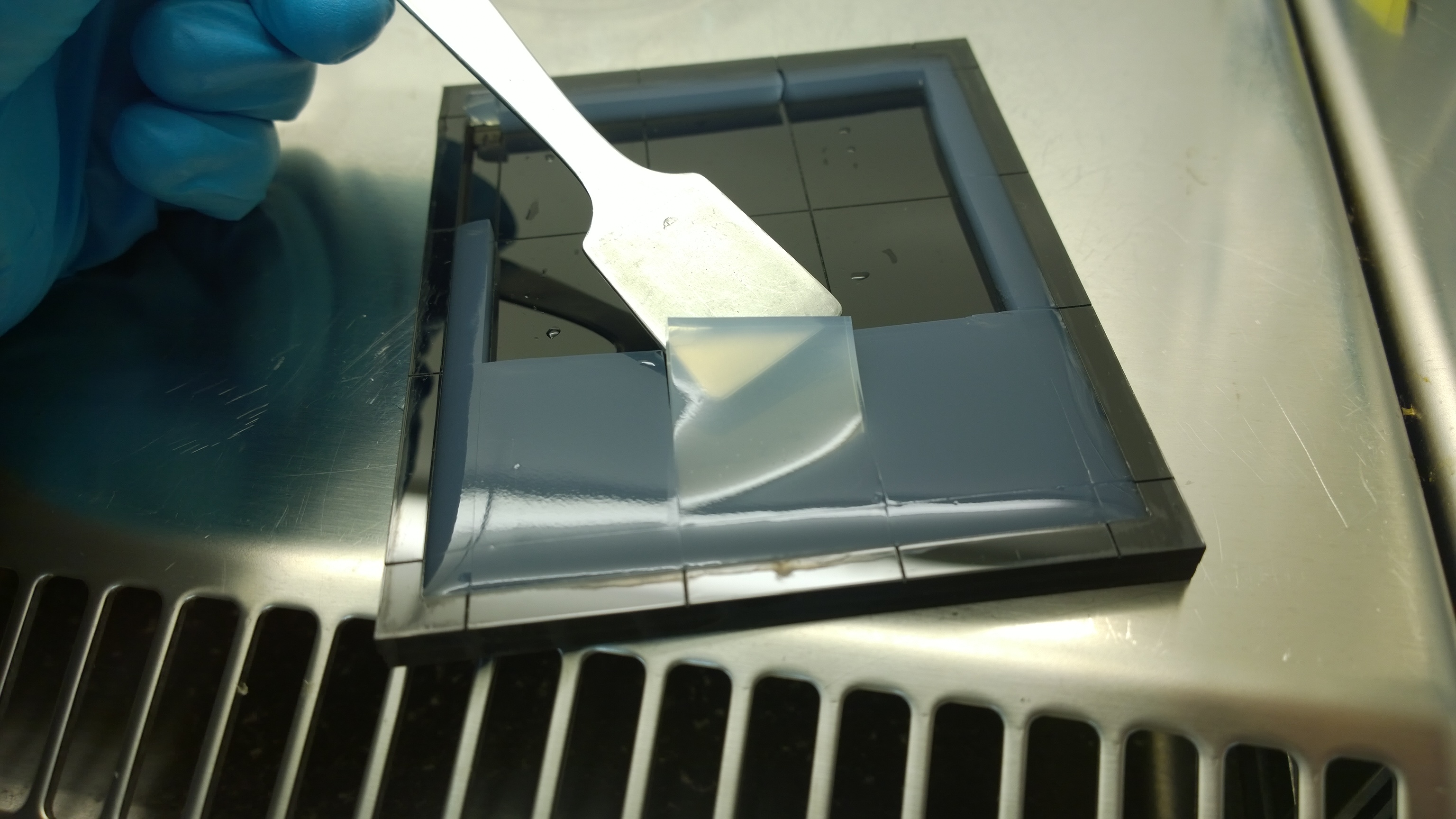
{{Team:Aachen/FigureFloat|Aachen_2_chipform.jpg|title=Sensor chip manufacturing using the closed mold|subtitle=When injecting the liquid agar into a closed mold we encounter problems due to frequent bubble formation.|left|width=500px}} | {{Team:Aachen/FigureFloat|Aachen_2_chipform.jpg|title=Sensor chip manufacturing using the closed mold|subtitle=When injecting the liquid agar into a closed mold we encounter problems due to frequent bubble formation.|left|width=500px}} | ||
| Line 151: | Line 148: | ||
{{Team:Aachen/FigureFloat|Aachen_Final_chipform.jpg|title=The finalized chip mold|subtitle=An open casting mold was found to be optimal for sensor chip manufacturing, because this approach was fast, easy to handle and generated a reproducible chip quality. In addition nine sensor chips could be produced simultaneously.|left|width=500px}} | {{Team:Aachen/FigureFloat|Aachen_Final_chipform.jpg|title=The finalized chip mold|subtitle=An open casting mold was found to be optimal for sensor chip manufacturing, because this approach was fast, easy to handle and generated a reproducible chip quality. In addition nine sensor chips could be produced simultaneously.|left|width=500px}} | ||
| - | |||
=== Induction of the sensor chips === | === Induction of the sensor chips === | ||
| - | |||
For initial testing of our molecular constructs, we simulated the presence of ''P. aeruginosa'' using IPTG or 3-oxo-C12 HSL. However, for induction a minimal volume is required as our initial experiments showed that diffusion of the inducers through the chip hindered the formation of distinct spots on the chips. An optimal and low volume of 0.2 µl was chosen for induction. Sensor cells based on ''E. coli'' BL21, which incorporated the [https://2014.igem.org/Team:Aachen/Parts#partsK1319042 K1319042] construct were able to detect IPTG concentrations down to 1 mM (0.2 µl), and as well for the sensor cells based on ''E. coli'' BL21 which incorporated the REACh constructs. Sensor cells based on ''E. coli'' BL21, which incorporated the [http://parts.igem.org/Part:BBa_K131026 K131026] construct were able to detect HSL concentrations down to 500 µg/ml (0.2 µl). Furthermore, detection of growing ''P. aeruginosa'' cells based on secreted HSLs was possible using the [http://parts.igem.org/Part:BBa_K131026 K131026] construct. The experiments conducted during induction of our sensor chips are described in more detail in the [https://2014.igem.org/Team:Aachen/Project/2D_Biosensor#biosensorachievements Achievements] section. | For initial testing of our molecular constructs, we simulated the presence of ''P. aeruginosa'' using IPTG or 3-oxo-C12 HSL. However, for induction a minimal volume is required as our initial experiments showed that diffusion of the inducers through the chip hindered the formation of distinct spots on the chips. An optimal and low volume of 0.2 µl was chosen for induction. Sensor cells based on ''E. coli'' BL21, which incorporated the [https://2014.igem.org/Team:Aachen/Parts#partsK1319042 K1319042] construct were able to detect IPTG concentrations down to 1 mM (0.2 µl), and as well for the sensor cells based on ''E. coli'' BL21 which incorporated the REACh constructs. Sensor cells based on ''E. coli'' BL21, which incorporated the [http://parts.igem.org/Part:BBa_K131026 K131026] construct were able to detect HSL concentrations down to 500 µg/ml (0.2 µl). Furthermore, detection of growing ''P. aeruginosa'' cells based on secreted HSLs was possible using the [http://parts.igem.org/Part:BBa_K131026 K131026] construct. The experiments conducted during induction of our sensor chips are described in more detail in the [https://2014.igem.org/Team:Aachen/Project/2D_Biosensor#biosensorachievements Achievements] section. | ||
| - | |||
{{Team:Aachen/BlockSeparator}} | {{Team:Aachen/BlockSeparator}} | ||
Revision as of 20:58, 17 October 2014
|
|
|
|
|
|
 "
"