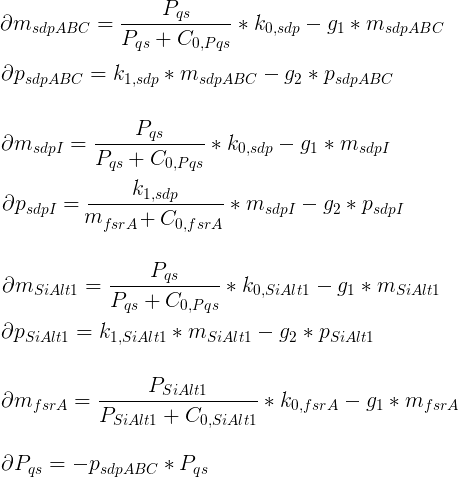
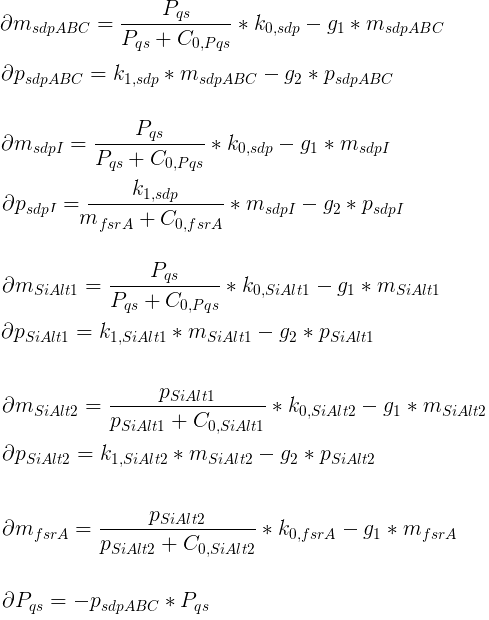
Variables
| msdpABC: | Quantity of SdpABC mRNA |
| psdpABC: | Quantity of SdpABC proteins |
| msdpI: | Quantity of SdpI mRNA |
| psdpI: | Quantity of SdpI proteins |
| mSiAlt1: | Quantity of sigma factor mRNA |
| pSiAlt1: | Quantity of sigma factor proteins |
| mfsrA: | Quantity of FsrA mRNA |
| pfsrA: | Quantity of FsrA proteins |
| Pqs: | Quantity of QS proteins |
Inquired Parameters
| C0,Pqs: | Basic concentration of QS proteins |
| C0,psdpI: | Basic concentration of SdpI proteins |
| C0,fsrA: | Basic concentration of FsrA proteins |
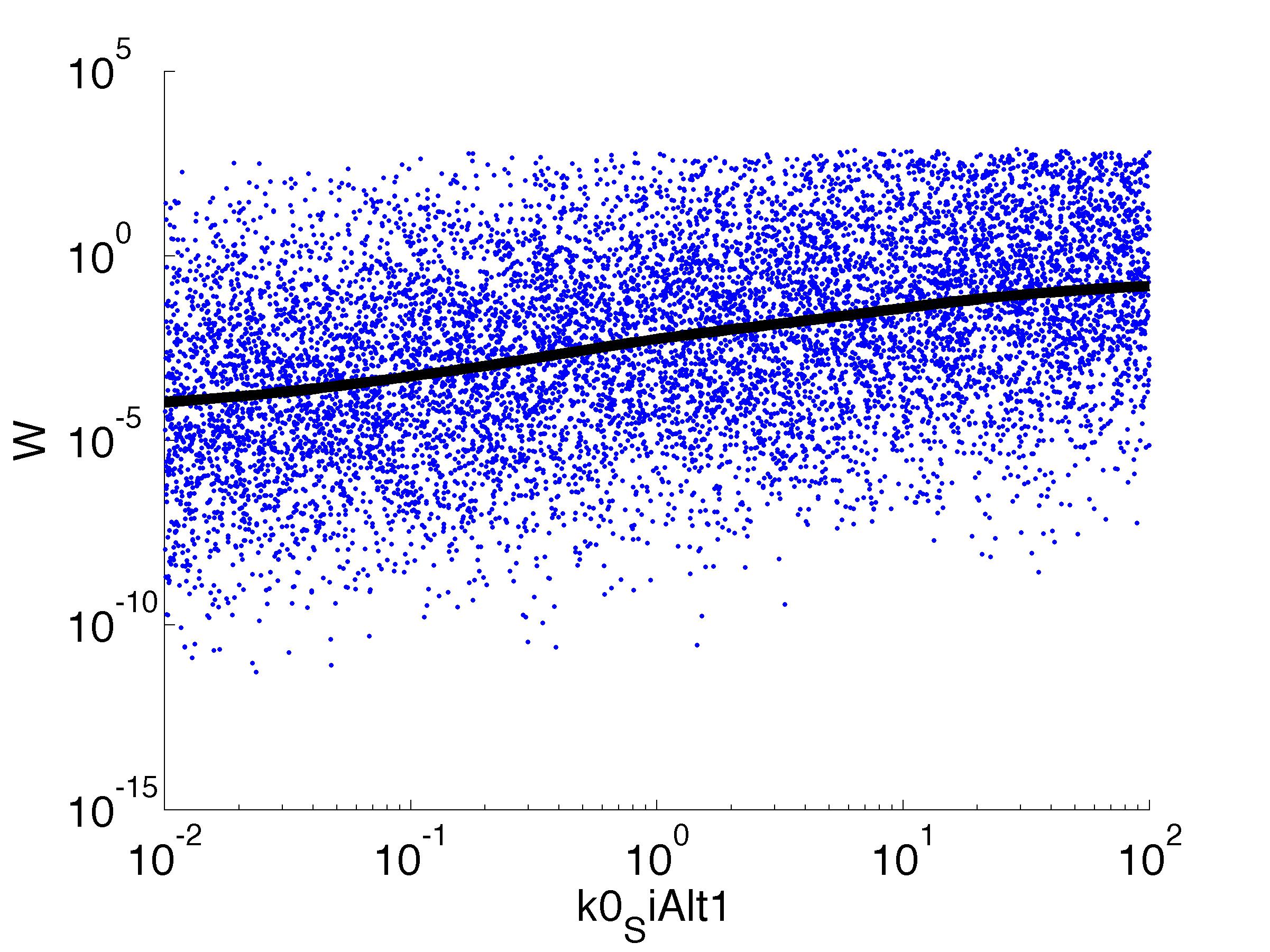
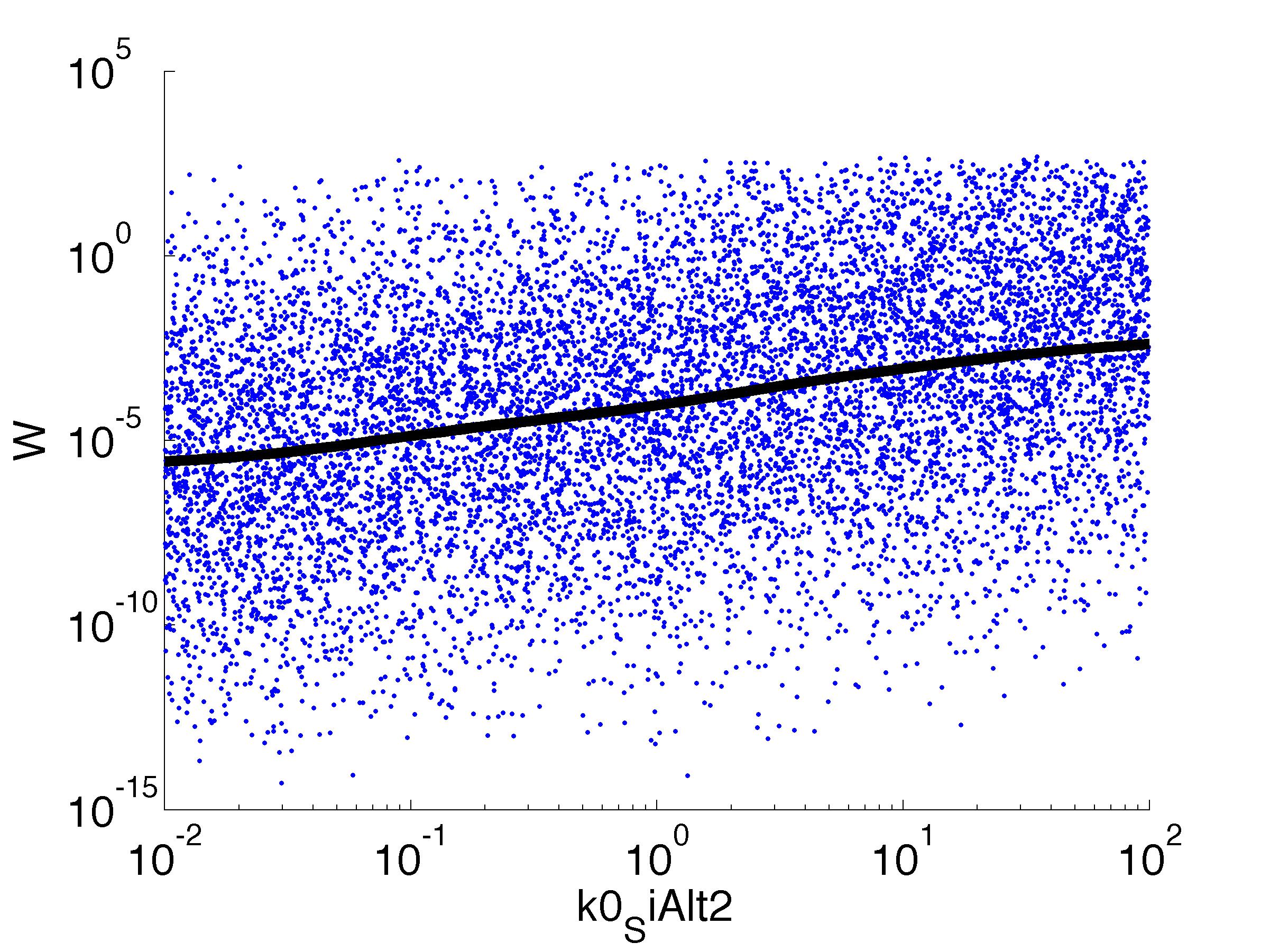
| k0,SiAlt1: | Transcription Rate of sigma factor |
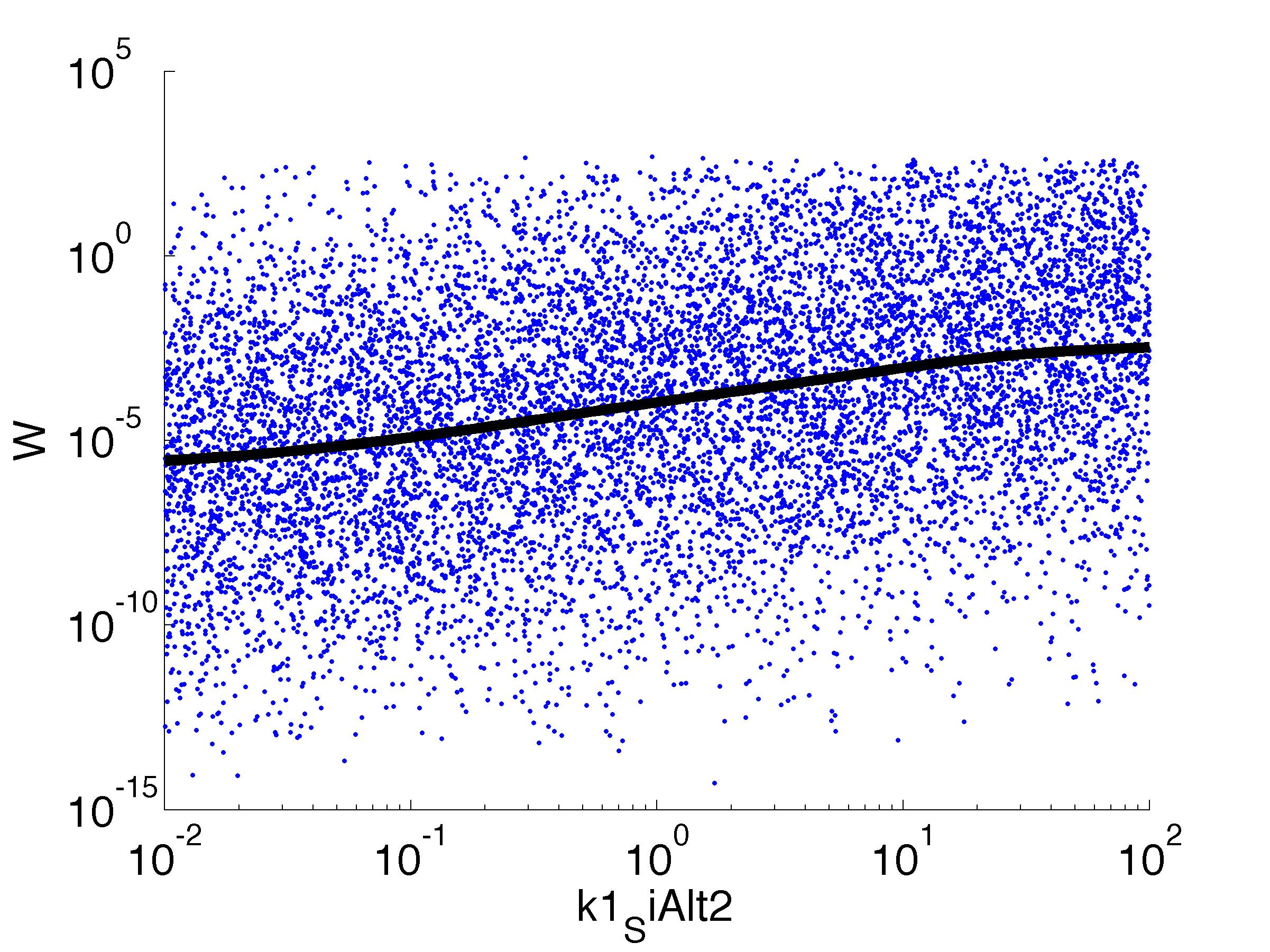
| k1,SiAlt1: | Translation Rate of sigma factor |
| C0,SiAlt1: | Basic concentration of sigma factor proteins |
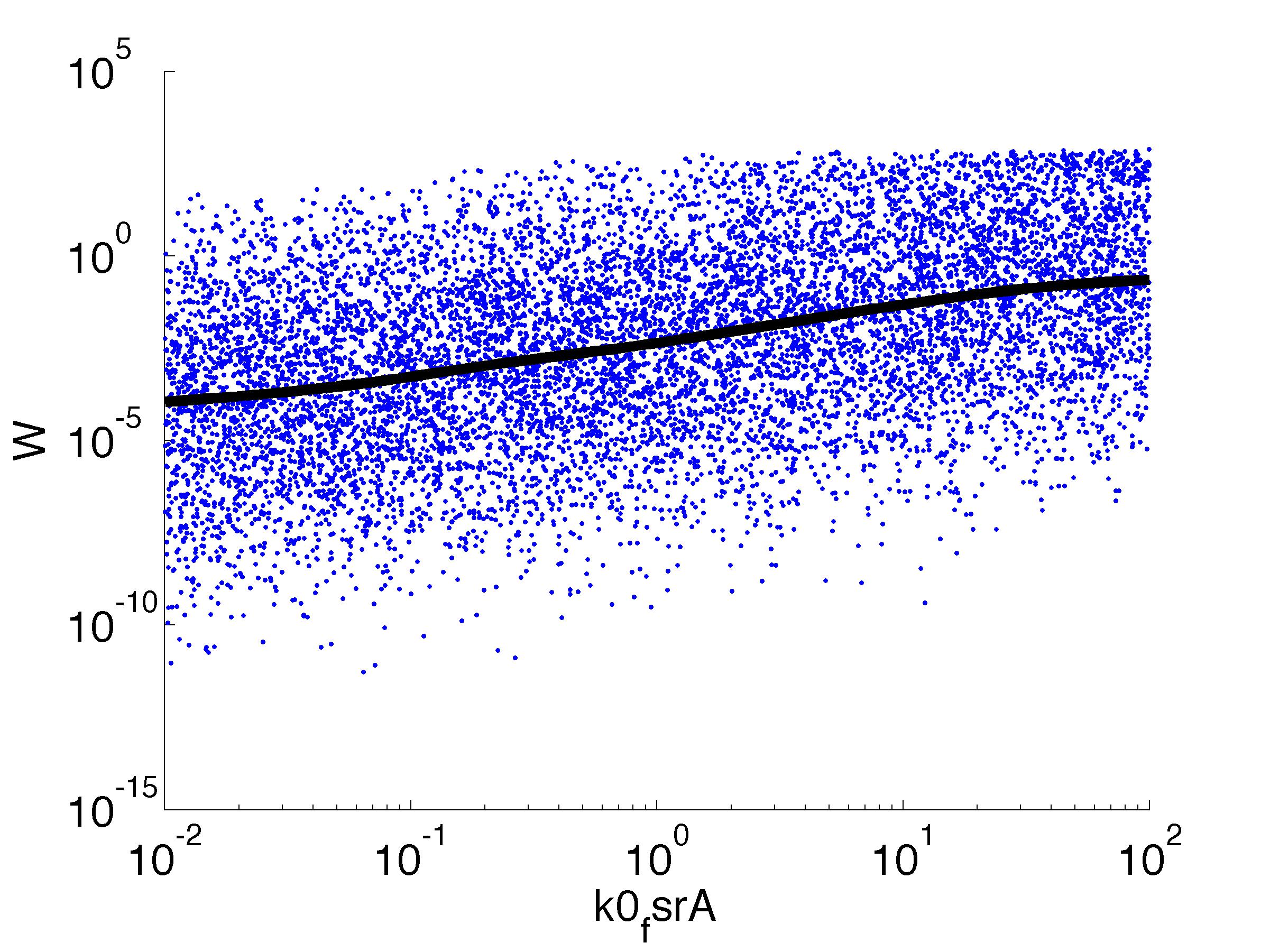
| k0,fsrA: | Transcription Rate of FsrA |
| k1,fsrA: | Translation Rate of FsrA |
Fixed Parameters
In a first evaluation, the variation of following parameters were figured out to cause only insignificant variation of the model's fitness. Therefore they were keept at these specified values for all subsequent simulations:
| Degradation rate (g1) of mRNA: | 12 molecules per hour |
| Degradation rate (g2) of proteins: | 1 molecule per hour |
| Transcription rate (k0) of Sdp: | 7.2 per hour |
| Translation rate (k1) of SdpI & SdpABC: | 10 per hour |
Fitness W
For analysis of the impact of each parameters, we used a function to determine the fitness W of the model. The fitness describes the integral of the surplus of toxic cannibalism proteins over the proprietary antidote. Therefore, a successful suicide equals a high fitness of the model. As we do not know the parameters in for our model we sampled the whole parameter space using a uniform sampler. Successively we used a Nadaraya-Watson kernel densiy estimator to approximate the expected value of W given a single parameter value. This allows us to asses the expected change in W when changing a single parameter, whithout knowledge of all other parameters.
 "
"