Team:Aachen/Notebook/Software/Measurarty
From 2014.igem.org
(→Statistical Region Merging (SRM)) |
|||
| Line 107: | Line 107: | ||
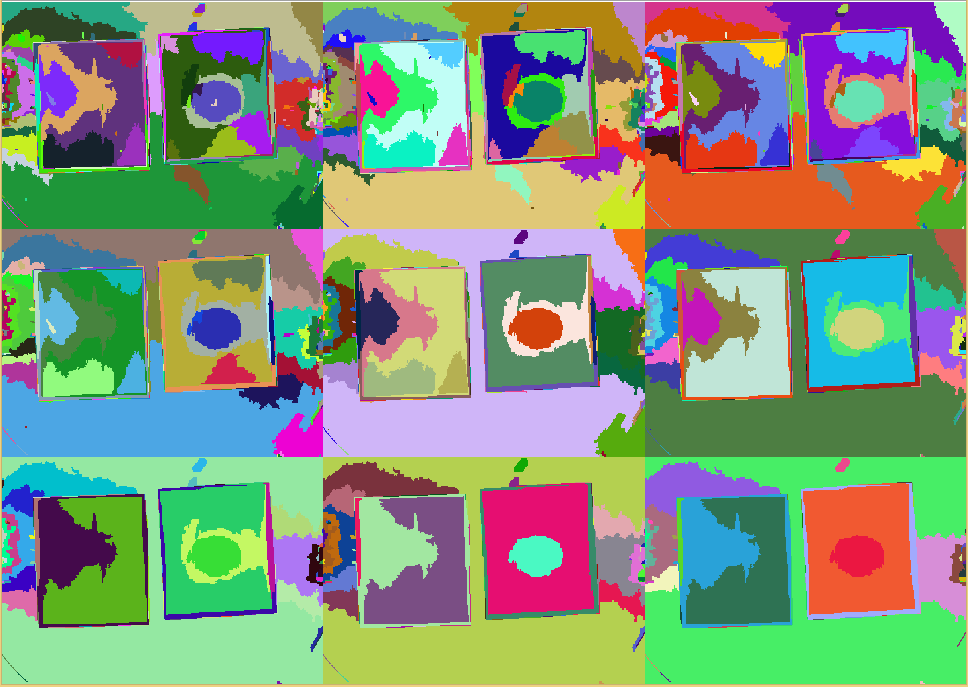
{{Team:Aachen/FigureDual|Aachen_srm_regions_3.PNG|Aachen_srm_regions_2.PNG|title1=SRM regions in random colors|title2=SRM regions (average color)|subtitle1=Different regions from an SRM run starting at $Q=256$ (top left) and going to $Q=1$ (bottom right). Each region is assigned a random color.|subtitle2=Different regions from an SRM run starting at $Q=256$ (top left) and going to $Q=1$ (bottom right). Each region is assigned the average color of that region.|width=425px}} | {{Team:Aachen/FigureDual|Aachen_srm_regions_3.PNG|Aachen_srm_regions_2.PNG|title1=SRM regions in random colors|title2=SRM regions (average color)|subtitle1=Different regions from an SRM run starting at $Q=256$ (top left) and going to $Q=1$ (bottom right). Each region is assigned a random color.|subtitle2=Different regions from an SRM run starting at $Q=256$ (top left) and going to $Q=1$ (bottom right). Each region is assigned the average color of that region.|width=425px}} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
=== SRM Clustering === | === SRM Clustering === | ||
| Line 139: | Line 134: | ||
<span class="anchor" id="segment"></span> | <span class="anchor" id="segment"></span> | ||
| - | In the segmentation stage all background regions | + | In the segmentation stage all background regions are removed. This task is quite crucial. If one removes too few, the final stage of finding pathogens might get irritated. |
| - | On the other hand, if one removes too many regions, positive hits might get removed early before detection. This surely | + | On the other hand, if one removes too many regions, positive hits might get removed early before detection. This surely must be avoided. |
We opted for a simple thresholding step because it showed that while being easy, it is an effective weapon against the uniform background. In fact, the good image quality we wanted to reach with our device allows now less sophisticated methods. | We opted for a simple thresholding step because it showed that while being easy, it is an effective weapon against the uniform background. In fact, the good image quality we wanted to reach with our device allows now less sophisticated methods. | ||
| Line 267: | Line 262: | ||
* link github | * link github | ||
| + | {{Team:Aachen/BlockSeparator}} | ||
| + | |||
| + | == References == | ||
| + | <span class="anchor" id="measurartyrefs"></span> | ||
| + | |||
| + | |||
| + | [1] Nock R, Nielsen F. Statistical region merging. IEEE Transactions on PAMI. 2004;26:1452–8. | ||
| + | |||
| + | [2] Boltz S. Statistical region merging matlab implementation; 2014. Available | ||
| + | from: [http://www.mathworks.com/matlabcentral/fileexchange/25619-image-segmentation-using-statistical-region-merging Matlab FileExchange] . Accessed 12 Dec 2013. | ||
{{Team:Aachen/Footer}} | {{Team:Aachen/Footer}} | ||
Revision as of 09:20, 17 October 2014
|
|
|
|
|
|
|
 "
"