Team:Aachen/Interlab Study
From 2014.igem.org
Aschechtel (Talk | contribs) (→Expected Results) |
Aschechtel (Talk | contribs) (→Constructs and strains) |
||
| Line 94: | Line 94: | ||
===Constructs and strains=== | ===Constructs and strains=== | ||
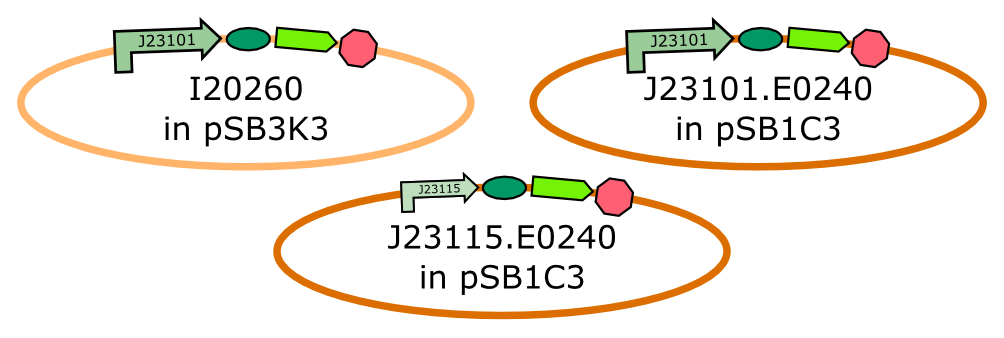
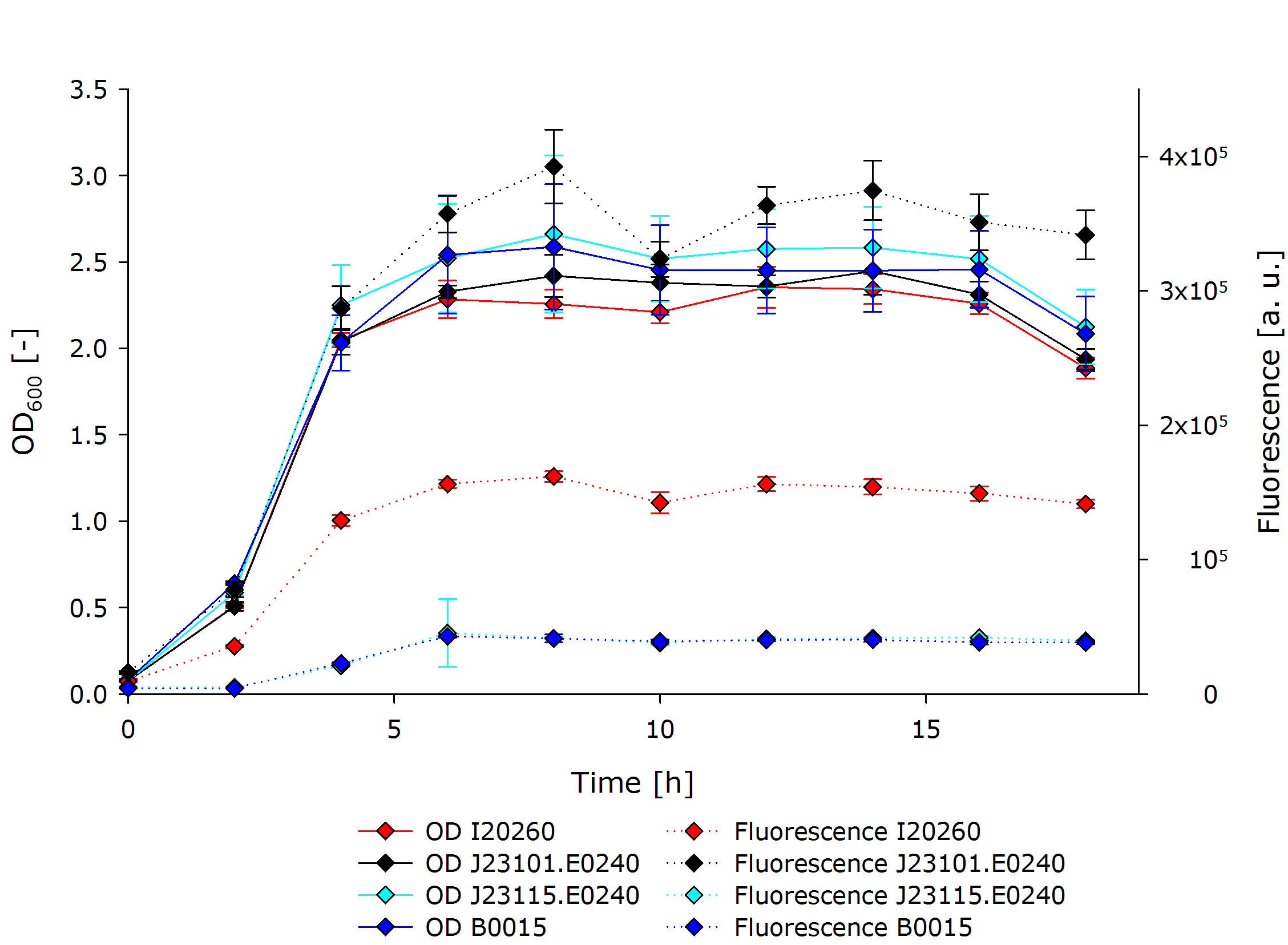
| - | All constructs used were transformed into [https://www.neb.com/products/c3019-neb-10-beta-competent-e-coli-high-efficiency NEB 10β] cells. The constructs I20260 as well as B0015 were taken directly from the iGEM 2014 distribution plates. The constructs J23101.E0240 as well as J23115.E0240 were made using the [http://parts.igem.org/Help:Assembly/3A_Assembly 3A Assembly]. Therefore, the subparts J23101, J23115 as well as E0240 were transformed directly from the 2014 distribution plates into [https://www.neb.com/products/c3019-neb-10-beta-competent-e-coli-high-efficiency NEB 10β] cells. Afterwards the plasmids were recovered using the [https://us.vwr-cmd.com/bin/public/demidoccdownload/50001659/7057R_ge_healthcare_illustra_nucleic_acid_sample_preparation.pdf illustra plasmidPrep Mini Spin Kit]. The purified plasmids J23101 and J23115 were cut with the restriction enzymes EcoRI and SpeI, while E0240 was cut with XbaI and PstI. The restricted plasmids were then ligated together using the T4 DNA Ligase. Afterwards, the ligation product was introduced into the pSB1C3 linearized backbone provided by iGEM headquarters with the 2014 distribution which we had also cut with EcoRI and PstI. All restrictions and ligations were performed using enzymes and buffers of the [http://shop2.neb-online.de/4DCGI/ezshop?action=Direktanzeige&Artikelnummer=NEBIGEM1%40&WorldNr=01&ButtonName=website&skontaktid=1055557&skontaktkey=RrbLvNPZxLRIFbyyyGOZambWfZKxFK NEB iGEM Kit] | + | All constructs used were transformed into [https://www.neb.com/products/c3019-neb-10-beta-competent-e-coli-high-efficiency NEB 10β] cells. The constructs I20260 as well as B0015 were taken directly from the iGEM 2014 distribution plates. The constructs J23101.E0240 as well as J23115.E0240 were made using the [http://parts.igem.org/Help:Assembly/3A_Assembly 3A Assembly]. Therefore, the subparts J23101, J23115 as well as E0240 were transformed directly from the 2014 distribution plates into [https://www.neb.com/products/c3019-neb-10-beta-competent-e-coli-high-efficiency NEB 10β] cells. Afterwards the plasmids were recovered using the [https://us.vwr-cmd.com/bin/public/demidoccdownload/50001659/7057R_ge_healthcare_illustra_nucleic_acid_sample_preparation.pdf illustra plasmidPrep Mini Spin Kit]. The purified plasmids J23101 and J23115 were cut with the restriction enzymes EcoRI and SpeI, while E0240 was cut with XbaI and PstI. The restricted plasmids were then ligated together using the T4 DNA Ligase. Afterwards, the ligation product was introduced into the pSB1C3 linearized backbone provided by iGEM headquarters with the 2014 distribution which we had also cut with EcoRI and PstI. All restrictions and ligations were performed using enzymes and buffers of the [http://shop2.neb-online.de/4DCGI/ezshop?action=Direktanzeige&Artikelnummer=NEBIGEM1%40&WorldNr=01&ButtonName=website&skontaktid=1055557&skontaktkey=RrbLvNPZxLRIFbyyyGOZambWfZKxFK NEB iGEM Kit]. The final product was once again transformed into [https://www.neb.com/products/c3019-neb-10-beta-competent-e-coli-high-efficiency NEB 10β] cells. |
The correct identity of the resulting constructs were confirmed by sequencing. The sequencing data (consensus sequences) can be found [https://2014.igem.org/File:Sequencing_Interlab_Study_iGEM_Aachen_2014.zip here]. | The correct identity of the resulting constructs were confirmed by sequencing. The sequencing data (consensus sequences) can be found [https://2014.igem.org/File:Sequencing_Interlab_Study_iGEM_Aachen_2014.zip here]. | ||
Revision as of 19:07, 16 October 2014
|
|
|
|
|
 "
"