Team:Aachen/Project/FRET Reporter
From 2014.igem.org
(→A Fluorescence Answer Faster Than Expression) |
(→A Fluorescence Answer Faster Than Expression) |
||
| Line 72: | Line 72: | ||
= A Fluorescence Answer Faster Than Expression = | = A Fluorescence Answer Faster Than Expression = | ||
<span class="anchor" id="fluorescence"></span> | <span class="anchor" id="fluorescence"></span> | ||
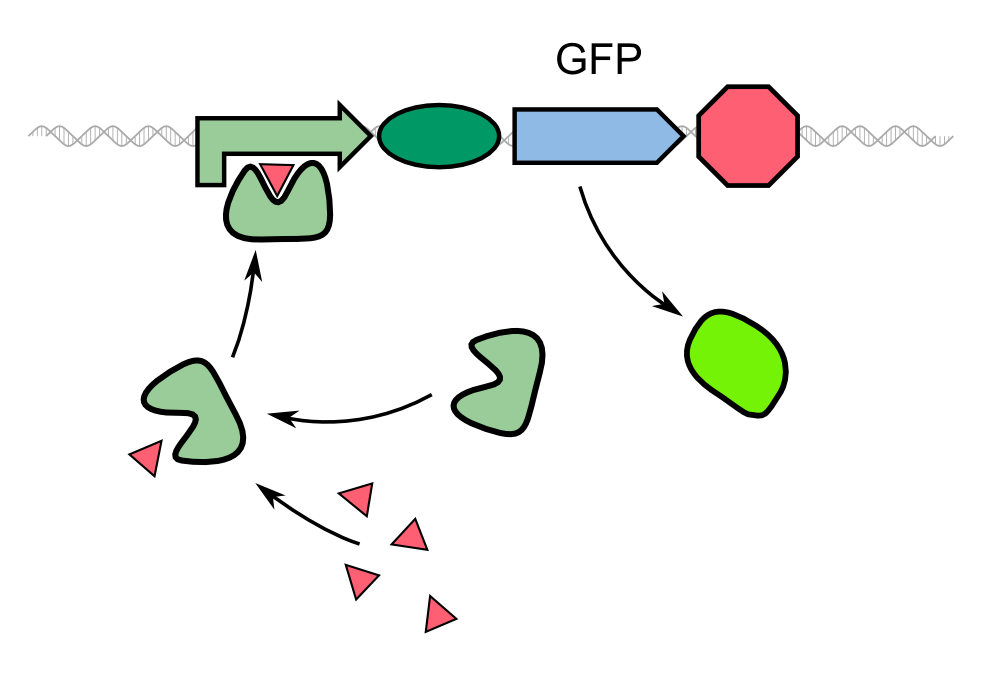
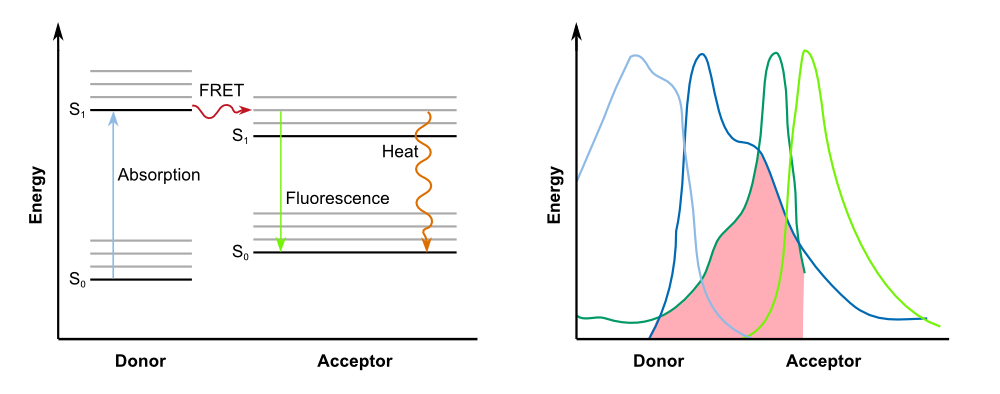
| - | Biosensors often work with a system that is comprised of a reported gene under the control of a promoter directly induced by the chemical that the sensor is supposed to detect. In the case of our 2D biosensor for ''Pseudomonas aeruginosa'', the expression of our reporter gene, GFP, would be directly induced by the activity of the bacterium's quorum sensing molecules. However, transcription, translation, folding and post-translational modifications take their time. Since our goal is to detect the pathogen as fast as possible, we wanted to use a system that gives a fluorescent answer fast than just expressing the fluorescent protein. | + | Biosensors often work with a system that is comprised of a reported gene under the control of a promoter directly induced by the chemical that the sensor is supposed to detect. In the case of our 2D biosensor for ''Pseudomonas aeruginosa'', the expression of our reporter gene, GFP, would be directly induced by the activity of the bacterium's quorum sensing molecules. However, transcription, translation, folding and post-translational modifications take their time. Since our goal is to detect the pathogen as fast as possible, we wanted to use a system that gives a fluorescent answer fast than just expressing the fluorescent protein. |
<center> | <center> | ||
| Line 78: | Line 78: | ||
</center> | </center> | ||
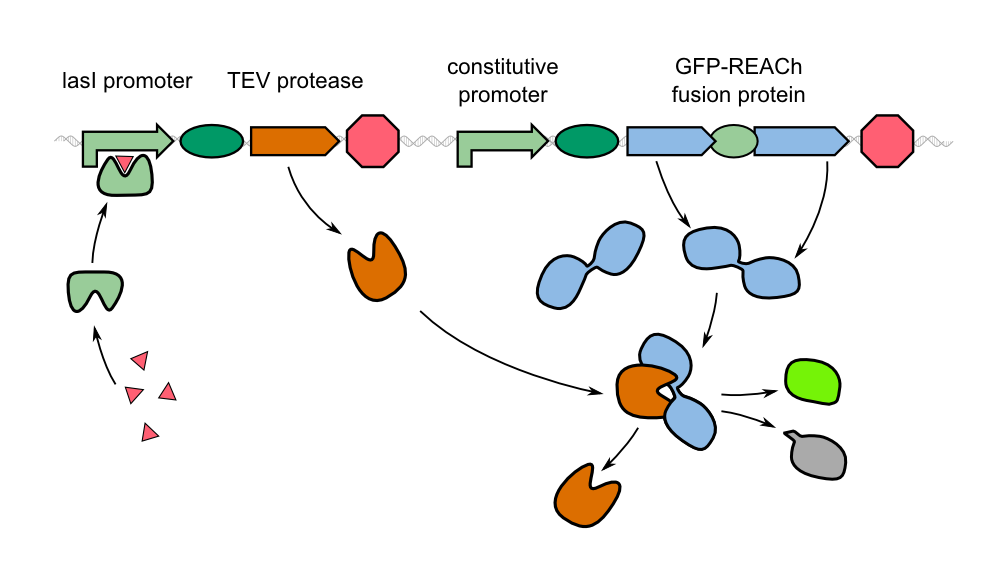
| - | Instead of the traditional approach, we '''constitutively express our reporter gene in a quenched form'''. As | + | Instead of the traditional approach, we '''constitutively express our reporter gene in a quenched form'''. As GFP-REACh fusion protein, fluorescence is suppressed. Our biosensor gives a response when homoserine lactones of ''Pseudomonas aeruginosa'' are taken up by our sensor cells where the '''autoinducer activates the expression of the TEV protease''' by binding to the LasI promoter in front of the protease gene. |
<center> | <center> | ||
| Line 85: | Line 85: | ||
This approach has two advantages: | This approach has two advantages: | ||
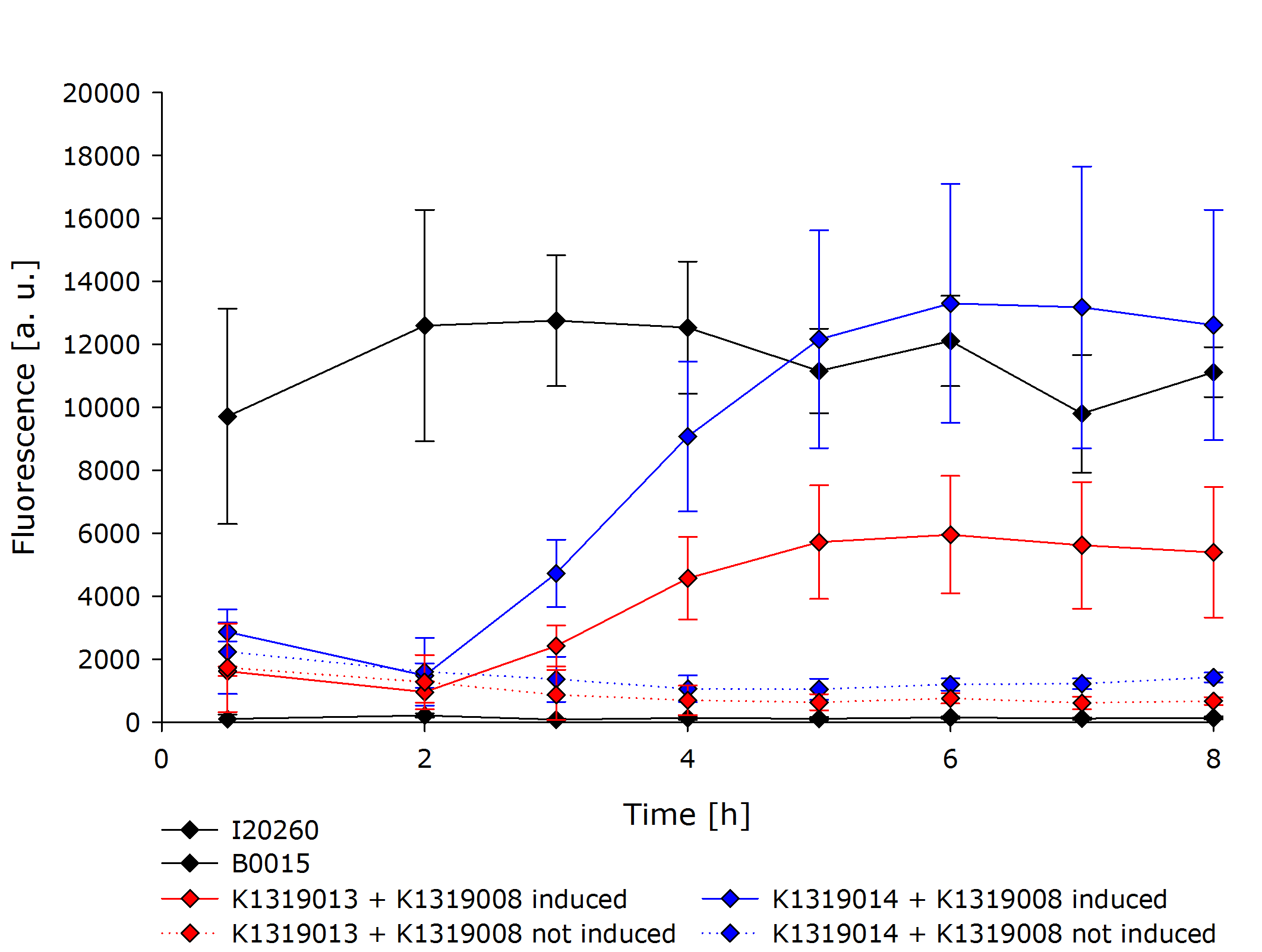
| - | * When ''Pseudomonas aeruginosa'' is detected by our cells, the reporter protein | + | * When ''Pseudomonas aeruginosa'' is detected by our cells, the reporter protein has already been expressed and only waits to be cleaved off the REACh quencher. The cleavage reaction catalyzed by the TEV protease is a faster process than expression and correct folding of GFP. This way we hope for '''an earlier response''' by our sensor cells. |
| - | * While a certain concentration of homoserine lactone will produce the same number of gene read-outs, one TEV protease can cleave many | + | * While a certain concentration of homoserine lactone will produce the same number of gene read-outs, one TEV protease can cleave many GFP-REACh constructs. Through the cleavage step, we introduce an '''amplification step''' into our system. With the TEV protease, we will be able to produce '''a much stronger signal''' in a short time interval. |
<center> | <center> | ||
| Line 93: | Line 93: | ||
</center> | </center> | ||
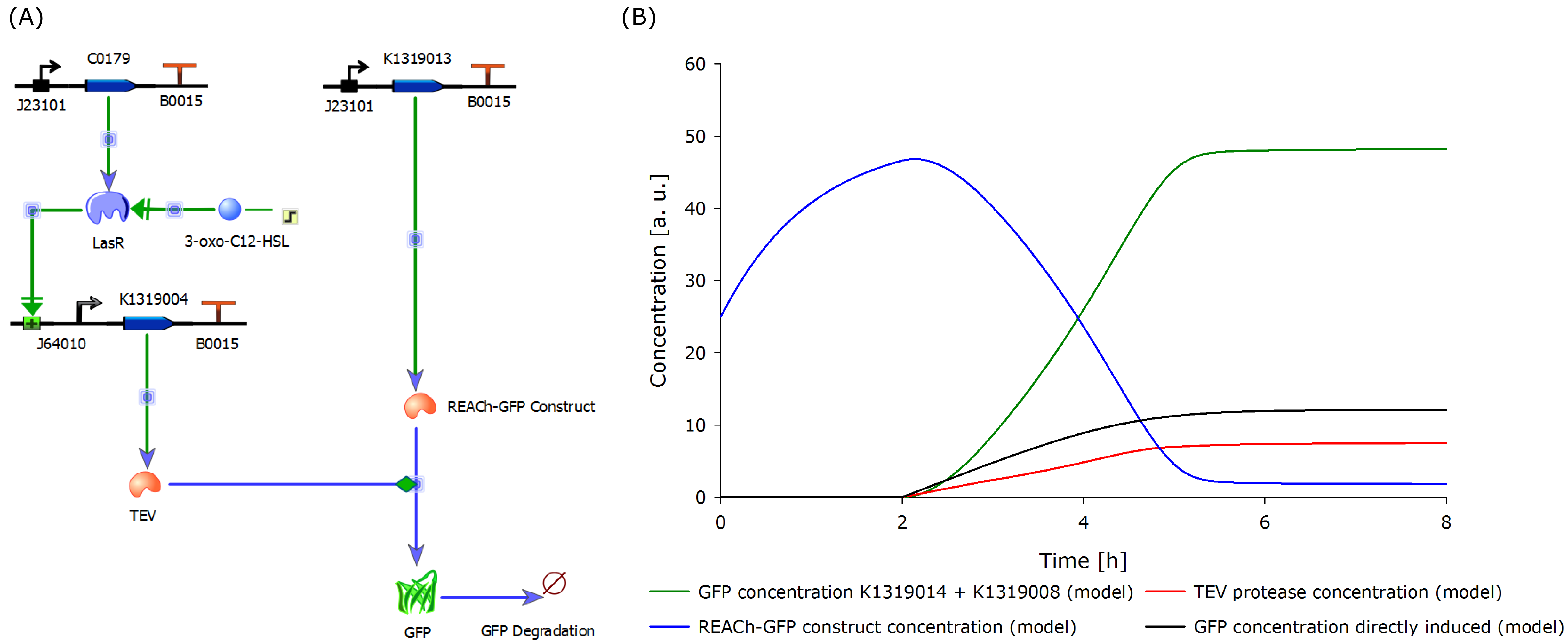
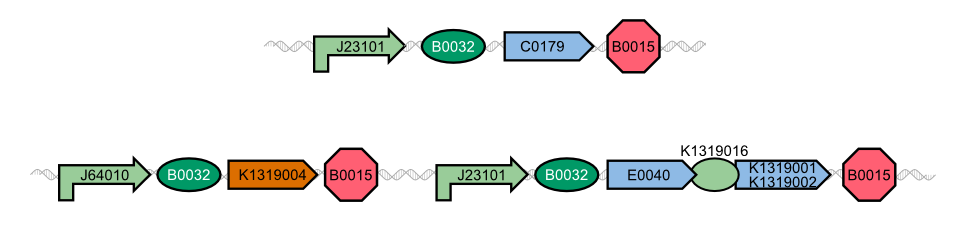
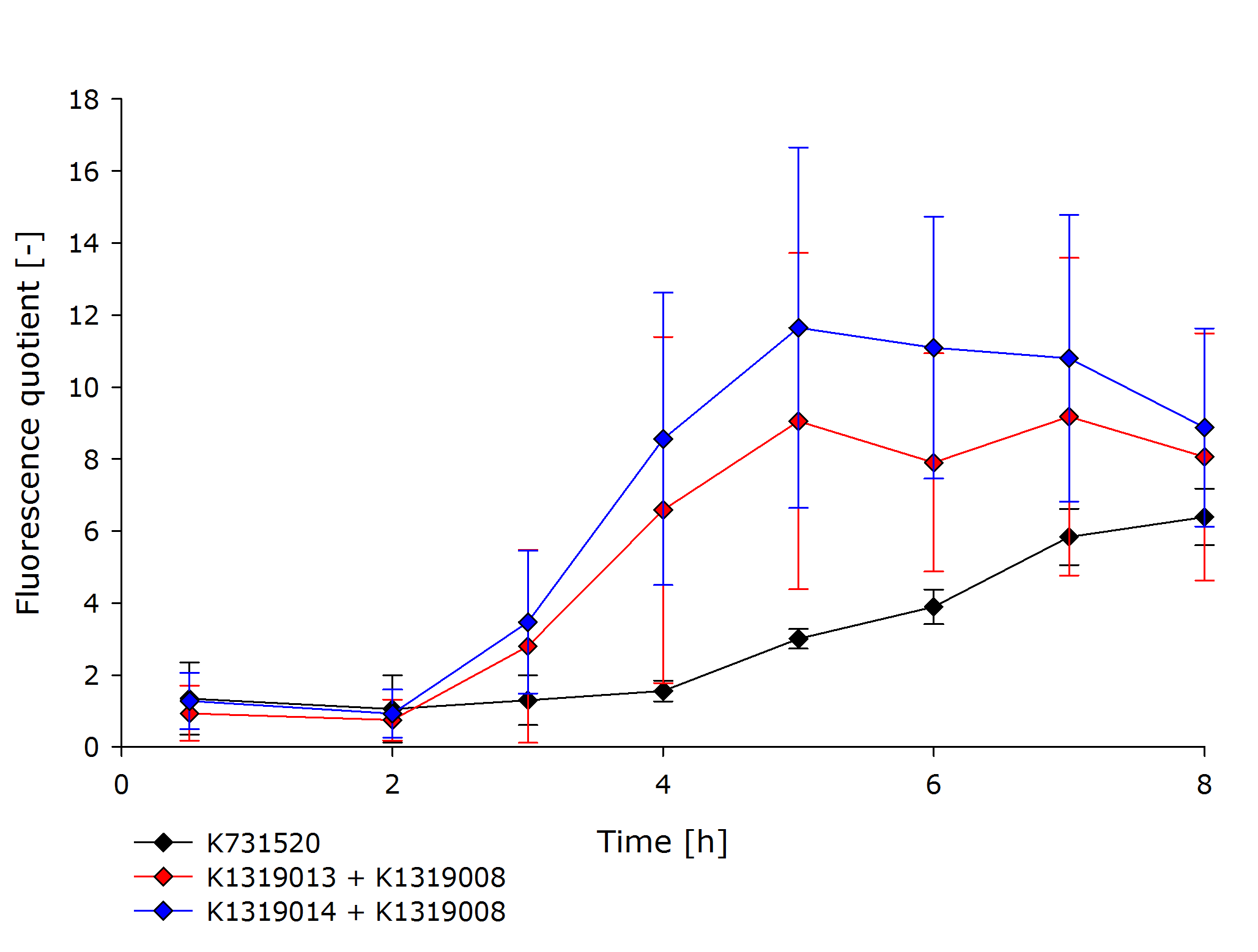
| - | The novel biosensor approach was modeled as shown above. The plotted results also include a model of a direct expression of GFP as it appears in traditional biosensors. The strength of the promotor used for the traditional approach is twice as high as the strength of the promotor upstream of the TEV coding sequence in | + | The novel biosensor approach was modeled as shown above. The plotted results also include a model of a direct expression of GFP as it appears in traditional biosensors. The strength of the promotor used for the traditional approach is twice as high as the strength of the promotor upstream of the TEV coding sequence in our novel approach. Despite the stronger promotor, a '''higher GFP concentration is generated in the model of the novel biosensor''', proving the stronger and faster output of our construct in theory. |
{{Team:Aachen/BlockSeparator}} | {{Team:Aachen/BlockSeparator}} | ||
Revision as of 18:55, 16 October 2014
|
|
|
|
|
|
|
|
 "
"