Team:Aachen/Project/Gal3
From 2014.igem.org
m |
|||
| Line 60: | Line 60: | ||
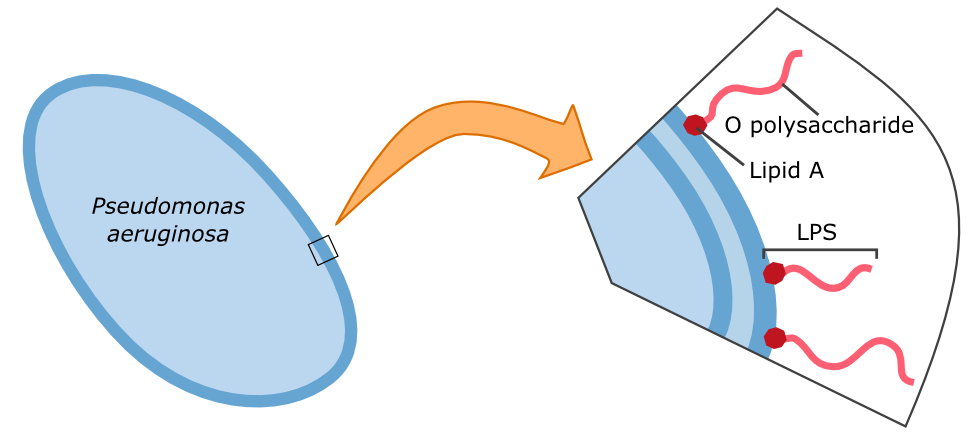
{{Team:Aachen/FigureFloat|Aachen_14-10-09_Pseudomonas_LPS_iNB.png|title=Cell wall composition of ''Pseudomonas aeruginosa''|subtitle=Gram-negative bacteria have two cell membranes. The LPS are embedded in the outer membrane and are composed of a lipid and an O polysaccharide.|width=420px}} | {{Team:Aachen/FigureFloat|Aachen_14-10-09_Pseudomonas_LPS_iNB.png|title=Cell wall composition of ''Pseudomonas aeruginosa''|subtitle=Gram-negative bacteria have two cell membranes. The LPS are embedded in the outer membrane and are composed of a lipid and an O polysaccharide.|width=420px}} | ||
| - | The specific binding of galectin-3 enables the construction of such a detection system. Parts of the '''lipopolysaccharide structure (LPS)''' of ''Pseudomonas aeruginosa'' can be bound by galectin-3. Specifically, the O polysaccharide (see figure on the left) of the LPS is recognized by galectin-3. A fusion protein of galectin-3 and a reporter protein, such as a fluorescent protein, can be built and applied in the detection of ''Pseudomonas aeruginosa''. | + | The specific binding of galectin-3 enables the construction of such a detection system. Parts of the '''lipopolysaccharide structure (LPS)''' of ''Pseudomonas aeruginosa'' can be bound by galectin-3. Specifically, the O polysaccharide (see figure on the left) of the LPS is recognized by galectin-3. A fusion protein of galectin-3 and a reporter protein, such as a fluorescent protein, can be built and applied in the detection of ''Pseudomonas aeruginosa''. |
| - | In our approach, a '''galectin-3-YFP fusion protein''' is built and expressed in ''E. coli''. A his-tag and a snap-tag for purification are included. The fusion protein can then be incorporated into a '''cell-free biosensor system'''. Such biosensors have many advantages over systems that use living cells; storage, for example, is much easier. From a [https://2014.igem.org/Team:Aachen/Safety biosafety] and social acceptance perspective, it is also advantageous if the sensor system does not contain live genetically modified organisms. | + | In our approach, a '''galectin-3-YFP fusion protein''' is built and expressed in ''E. coli''. A his-tag and a snap-tag for purification are included. The fusion protein can then be incorporated into a '''cell-free biosensor system'''. Such biosensors have many advantages over systems that use living cells; storage, for example, is much easier. From a [https://2014.igem.org/Team:Aachen/Safety biosafety] and social acceptance perspective, it is also advantageous if the sensor system does not contain live genetically modified organisms. |
{{Team:Aachen/FigureFloatRight|align=center|Aachen_14-10-09_Cell_Free_Biosensor_iNB.png|width=500px}} | {{Team:Aachen/FigureFloatRight|align=center|Aachen_14-10-09_Cell_Free_Biosensor_iNB.png|width=500px}} | ||
| Line 70: | Line 70: | ||
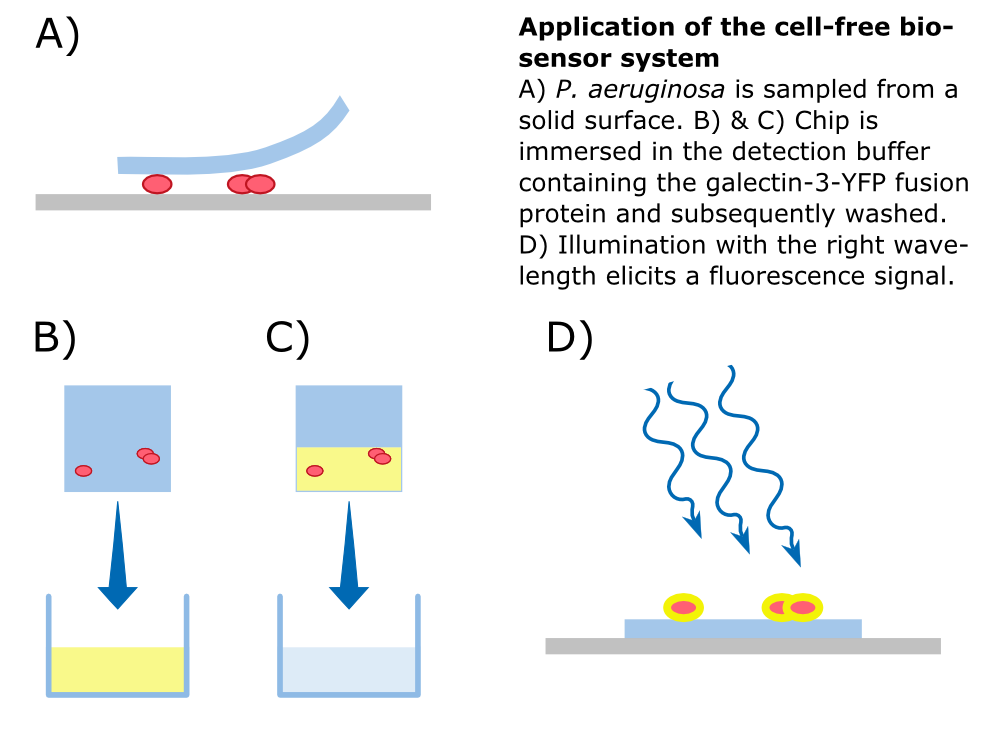
| - | To detect ''P. aeruginosa'' cells, an agar chip could be used to sample a solid surface. However, other materials but agar can be considered to collect the pathogens. The cell stick to the sampling chip which is then immersed in a detection buffer containing the galectin-3-YFP fusion protein. Excess protein is removed during washing in a suitable buffer. The galectin-3 remains bound to the pathogen and '''illumination with 514 nm''', the excitation frequency of YFP, in a modified version of our measurement device reveals the location of the cells. The picture taken by the measurement device can then be analyzed by our software ''Measurarty''. | + | To detect ''P. aeruginosa'' cells, an agar chip could be used to sample a solid surface. However, other materials but agar can be considered to collect the pathogens. The cell stick to the sampling chip which is then immersed in a detection buffer containing the galectin-3-YFP fusion protein. Excess protein is removed during washing in a suitable buffer. The galectin-3 remains bound to the pathogen and '''illumination with 514 nm''', the excitation frequency of YFP, in a modified version of our measurement device reveals the location of the cells. The picture taken by the measurement device can then be analyzed by our software ''Measurarty''. |
{{Team:Aachen/BlockSeparator}} | {{Team:Aachen/BlockSeparator}} | ||
| Line 79: | Line 79: | ||
<span class="anchor" id="naturalfunctions"></span> | <span class="anchor" id="naturalfunctions"></span> | ||
| - | Galectins are proteins of the lectin family, which ''' | + | Galectins are proteins of the lectin family, which posess '''carbonhydrate recognition domains''' binding specifically to β-galactoside sugar residues. In humans, 10 different galectines have been identified, among which is galectin-3. |
Galectin-3 has a size of about 31 kDA and is encoded by a single gene, LGALS3. It has many physiological functions, such as cell adhesion, cell growth and differentiation, and contributes to the development of cancer, inflammation, fibrosis and others. | Galectin-3 has a size of about 31 kDA and is encoded by a single gene, LGALS3. It has many physiological functions, such as cell adhesion, cell growth and differentiation, and contributes to the development of cancer, inflammation, fibrosis and others. | ||
Human galectin-3 is a protein of the lectin-family that was shown to bind the LPS of multiple human pathogens. | Human galectin-3 is a protein of the lectin-family that was shown to bind the LPS of multiple human pathogens. | ||
| - | Some of them, including '' | + | Some of them, including ''Pseudomonas aeruginosa'' protect themselves against the human immune system by mimicking the lipopolysaccharides (LPS) present on human erythrocytes. |
By making fusion proteins of galectin-3 with fluorescent reporter proteins, pathogens can be labelled and made visible by fluorescence-microscopy. | By making fusion proteins of galectin-3 with fluorescent reporter proteins, pathogens can be labelled and made visible by fluorescence-microscopy. | ||
| Line 95: | Line 95: | ||
<span class="anchor" id="gal3achievements"></span> | <span class="anchor" id="gal3achievements"></span> | ||
| - | + | Due to the generous support of Sophia Böcker and Prof. Elling of the Helmholtz Institute for Biomedical Engineering, we got access to an pET17-derived expression plasmid for a His- and SNAP-tagged YFP-galectin-3 fusion protein. We transformed the fusion protein into ''E. coli'' Rosetta cells and conducted a batch fermentation to obtain large amounts of protein (FIGURE). | |
| + | |||
| + | |||
| + | |||
| + | With the help of David Schönauer and Alan Mertens from the Institute for Biotechnolgy we then purified the fusion protein using by FPLC. | ||
| + | |||
| + | |||
| + | |||
| + | We attempted to do an experiment to test the binding of the Gal-3 fusion protein to the LPS of [TARGET_ORGANISMS] as shown previously [LITERATURE-REFERENCE], but we were unable to obtain useful results because our fluorescence microscope was not sensitive enough. | ||
| + | |||
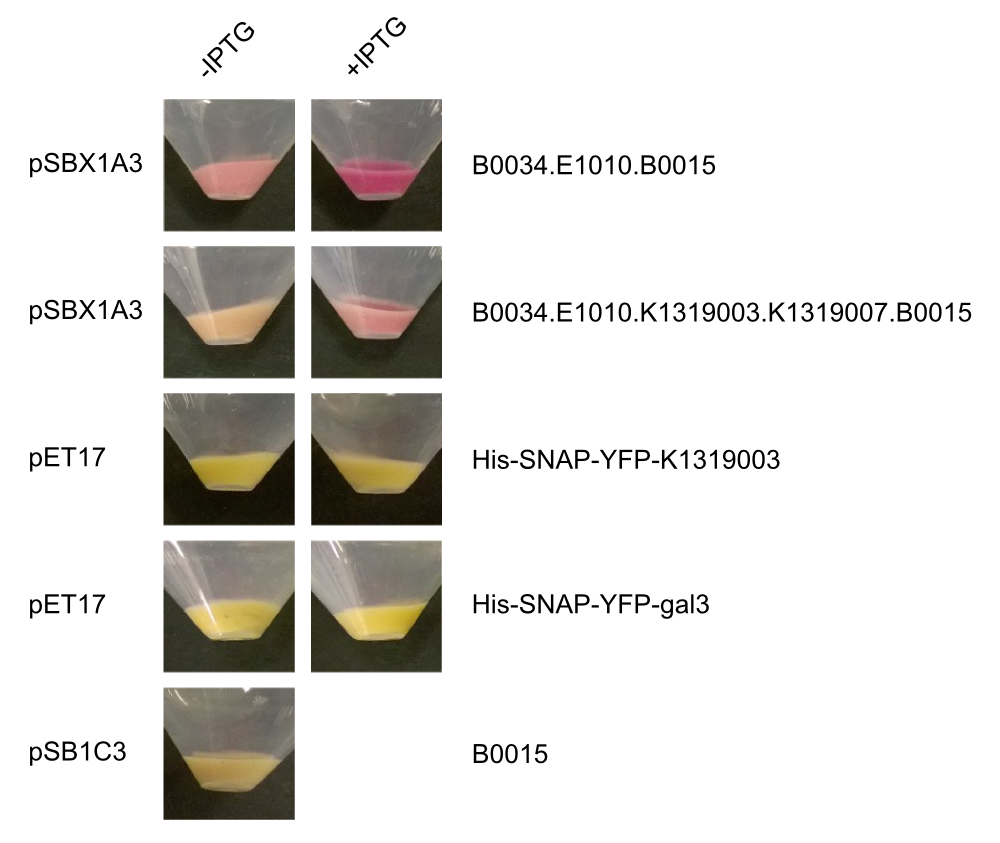
| + | After we received the collection of pSBX-expression vectors from Team Heidelberg, we used Gibson assembly to make K1319020 from K1319003 and pSBX1A3, which is the translational unit for a mRFP-Gal3 fusion protein with a C-terminal 6xHis tag: | ||
| + | <center>{{Team:Aachen/Figure|File:Aachen_K1319020.png|width=500px|title=K1319009|This BioBrick is a construction intermediate of K1319003 (gal3), E1010 (mRFP), K1319007 (6xHis tag) to K1319020 (translational unit of the fusion protein).}}</center> | ||
| + | |||
| + | We also cloned our K1319003 into the pET17 expression vector and expressed all combinations of fusion proteins in E. coli BL21(DE3). A SDS-PAGE showed that all fusion proteins were fully translated: | ||
| + | |||
| + | {{Team:Aachen/FigureFloat|File:Aachen_Gal3_Expression.png|width=350px|title=SDS-PAGE of K1319020 expression|subtitle=The fusion protein was fully translated to the correct molecular mass of 74 kDa.}} | ||
| + | |||
| - | |||
<center> | <center> | ||
Revision as of 15:41, 16 October 2014
|
|
|
|
 "
"